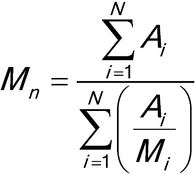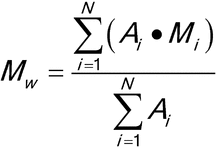Chitosan
DEFINITION
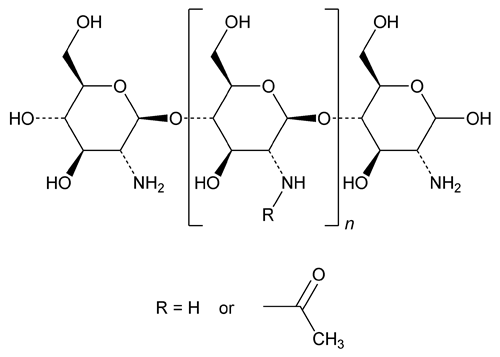
Chitosan is an unbranched binary polysaccharide consisting of N-acetyl-d-glucosamine and d-glucosamine units linked in a  (1®4) manner. Chitosan is obtained by partial deacetylation of chitin, which is extracted from the shells of edible shrimps and crabs suitable for human use. Its degree of deacetylation is NLT 70.0% and NMT 95.0%.
(1®4) manner. Chitosan is obtained by partial deacetylation of chitin, which is extracted from the shells of edible shrimps and crabs suitable for human use. Its degree of deacetylation is NLT 70.0% and NMT 95.0%.
IDENTIFICATION
• B.
Sample:
0.2 g of Chitosan powder
Analysis:
Add 80 g of water to the Sample, and stir briefly to obtain a dispersion. Separately prepare a glycolic acid solution by dissolving 0.1 g of glycolic acid in 20 g of water. Add the solution in one step to the dispersion. Stir the mixture gently at room temperature until it becomes a clear solution. [Note—It takes approximately 30–60 min to obtain a clear solution. ] Add 5 g of a 0.5% sodium lauryl sulfate aqueous solution to the clear solution.
Acceptance criteria:
A gelatinous mass is formed.
ASSAY
• Degree of Deacetylation
[Note—If tetramethylsilane is not used as the NMR reference, a suitable signal of the solvent itself can be used as a reference. ]
Solvent:
Deuterated formic acid
NMR reference:
Tetramethylsilane
Sample solution:
Into a 20-mL scintillation vial with a screw cap, dissolve 5–10 mg of Chitosan in deuterated formic acid, containing 0.5%–1.0% of tetramethylsilane, to obtain 1 mL of solution. Tightly close the vial, and dissolve Chitosan using a magnetic stirrer. To completely dissolve takes it about 48 h; the stirring is stopped when a clear solution with a high viscosity is obtained. Break up any clumps formed during the dissolution process with a spatula, if necessary.
Analysis
Sample:
Sample solution
Transfer 0.5–1.0 mL of the Sample solution to a standard 5-mm NMR spinning tube. Proceed as directed in Nuclear Magnetic Resonance  761
761 , Relative Method of Quantitation, scanning the region from 0–7 ppm, and using the calculation formulas below. Record as A1 the average area of the composite band from about 6–3 ppm, representing the seven protons with oxygen neighbors in the sugar ring. Record as A2 the average area of the signals at about 2 ppm, due to the methyl groups of the acetyl units, with reference to the tetramethylsilane singlet at 0 ppm.
, Relative Method of Quantitation, scanning the region from 0–7 ppm, and using the calculation formulas below. Record as A1 the average area of the composite band from about 6–3 ppm, representing the seven protons with oxygen neighbors in the sugar ring. Record as A2 the average area of the signals at about 2 ppm, due to the methyl groups of the acetyl units, with reference to the tetramethylsilane singlet at 0 ppm.
Calculate the percentage of deacetylation degree, by weight, in the Chitosan taken:
Result = {1  [(7 × A2)/(3 × A1)] × 100
[(7 × A2)/(3 × A1)] × 100
Acceptance criteria:
The degree of deacetylation is NLT 70.0% and NMT 95.0%.
IMPURITIES
• Residue on Ignition  281
281 :
NMT 1.0%
:
NMT 1.0%
• Heavy Metals, Method III  231
231 :
NMT 10 ppm
:
NMT 10 ppm
• Limit of Lead, Mercury, Chromium, Nickel, Cadmium, and Arsenic
65% Nitric acid:
Use ultratrace nitric acid, 65%–70% HNO3, ACS reagent grade.
Internal standard:
Transfer 0.2 mL of a solution containing 1000 ppm of yttrium [Note—Yttrium ICP standard solutions are commercially available.1 ] and 0.2 mL of a solution containing 1000 ppm of lutetium [Note—Lutetium ICP standard solutions are commercially available.2 ] to a 100-mL volumetric flask, add 1 mL of 65% Nitric acid, dilute with water to volume, and mix.
Blank standard:
Transfer 1.0 mL of the Internal standard to a 100-mL volumetric flask, add 1 mL of 65% Nitric acid, dilute with water to volume, and mix.
Standard solutions:
Transfer 0.1 mL of a solution containing 10 ppm of each of lead, mercury, chromium, nickel, cadmium, and arsenic [Note—Multi-element ICP standard solutions are commercially available.3 ] to a 100-mL volumetric flask, add 1 mL of the Internal standard and 1 mL of 65% Nitric acid, and dilute with water to volume (Standard solution 0.01 ppm). Transfer 0.1 mL of a solution containing 10 ppm of each of lead, mercury, chromium, nickel, cadmium, and arsenic to a 1000-mL volumetric flask, add 10 mL of the Internal standard and 10 mL of 65% Nitric acid, and dilute with water to volume (Standard solution 0.001 ppm).
Sample solution:
Transfer 1.0 g of Chitosan to a clean, dry, 100-mL Kjeldahl flask. [Note—A 300-mL flask may be used if the reaction foams excessively. ] Clamp the flask at an angle of 45 , and add a sufficient quantity of a mixture of 8 mL of sulfuric acid and 10 mL of nitric acid to moisten the substance thoroughly. Warm gently until the reaction commences, allow the reaction to subside, and add portions of the same acid mixture, heating after each addition, until a total of 18 mL of the acid mixture has been added. Increase the amount of heat, and boil gently until the solution darkens. Cool, add 2 mL of nitric acid, and heat again until the solution darkens. Continue the heating, followed by addition of nitric acid until no further darkening occurs, then heat strongly to the production of dense, white fumes. Cool, cautiously add 5 mL of water, boil gently to the production of dense, white fumes, and continue heating until the volume is reduced to a few mL. Cool, cautiously add 5 mL of water, and examine the color of the solution. If the color is yellow, cautiously add 1 mL of 30% hydrogen peroxide, and again evaporate to the production of dense, white fumes and a volume of 2–3 mL. If the solution is still yellow, repeat the addition of 5 mL of water and the peroxide treatment. Cool, dilute cautiously with a few mL of water, rinse into a 20-mL volumetric flask, and dilute with water to volume. Transfer 5.0 mL of digestion solution to a 100-mL volumetric flask, and retain the remaining digestion solution for use in the Limit of Iron. Add 1 mL of the Internal standard to the 100-mL volumetric flask, dilute with water to volume, and mix.
, and add a sufficient quantity of a mixture of 8 mL of sulfuric acid and 10 mL of nitric acid to moisten the substance thoroughly. Warm gently until the reaction commences, allow the reaction to subside, and add portions of the same acid mixture, heating after each addition, until a total of 18 mL of the acid mixture has been added. Increase the amount of heat, and boil gently until the solution darkens. Cool, add 2 mL of nitric acid, and heat again until the solution darkens. Continue the heating, followed by addition of nitric acid until no further darkening occurs, then heat strongly to the production of dense, white fumes. Cool, cautiously add 5 mL of water, boil gently to the production of dense, white fumes, and continue heating until the volume is reduced to a few mL. Cool, cautiously add 5 mL of water, and examine the color of the solution. If the color is yellow, cautiously add 1 mL of 30% hydrogen peroxide, and again evaporate to the production of dense, white fumes and a volume of 2–3 mL. If the solution is still yellow, repeat the addition of 5 mL of water and the peroxide treatment. Cool, dilute cautiously with a few mL of water, rinse into a 20-mL volumetric flask, and dilute with water to volume. Transfer 5.0 mL of digestion solution to a 100-mL volumetric flask, and retain the remaining digestion solution for use in the Limit of Iron. Add 1 mL of the Internal standard to the 100-mL volumetric flask, dilute with water to volume, and mix.
Blank solution:
Prepare the blank digestion solution, following the preparation procedure for the Sample solution, but without using Chitosan. Transfer 5.0 mL of blank digestion solution to a 100-mL volumetric flask, and retain the remaining digestion solution for use in the Limit of Iron. Add 1 mL of the Internal standard to the 100-mL volumetric flask, dilute with water to volume, and mix.
Instrumental conditions
(See Plasma Spectrochemistry  730
730 .)
.)
Mode:
Inductively coupled plasma–mass spectrometer (ICP–MS)
Spectrometer:
Quadrupole mass spectrometer
Detector:
Ion detector maintained under vacuum
Other requirements:
The instrument should read all isotopes for the following elements shown in Table 1, for the yttrium internal standard (89 amu) and the lutetium internal standard (175 and 176 amu), and should report the total element contents using the most naturally abundant isotopes.
Table 1
| Element | Isotope (amu) |
|---|---|
| Lead | 208 |
| Mercury | 201 |
| 202 | |
| Chromium | 53 |
| Nickel | 58 |
| 60 | |
| Cadmium | 114 |
| Arsenic | 75 |
Analysis
Samples:
Blank standard, Standard solutions, Sample solution, and Blank solution
Instrument performance must be verified to conform to the manufacturer’s specifications for resolution and sensitivity. Before analyzing samples, the instrument must pass a suitable performance check. Perform the evaluation using instrument software such as correction equations for interferences and taking the Internal standard into account. Generate the calibration curve using the Blank standard, Standard solution 0.001 ppm, and Standard solution 0.01 ppm: the linear regression coefficient is NLT 0.999.
Aspirate the Blank solution and Sample solution, respectively, at least in duplicate, and report the average reading as each element content of the sample. Determine the concentration CB, in µg/mL, of each element in the Blank solution, and also determine the concentration CS, in µg/mL, of each element in the Sample solution using the calibration curve.
Calculate the quantity, in µg/g, of each element in the portion of Chitosan taken:
Result = [(CS  CB)/W] × F × V
CB)/W] × F × V
| F | = | = dilution factor, 4 |
| V | = | = volume of the Sample solution, 100 mL |
| W | = | = weight of the Chitosan taken to prepare the Sample solution (g) |
Acceptance criteria:
See Table 2.
Table 2
| Element | Acceptance Criteria (ppm) |
|---|---|
| Lead | NMT 0.5 |
| Mercury | NMT 0.2 |
| Chromium | NMT 1.0 |
| Nickel | NMT 1.0 |
| Cadmium | NMT 0.2 |
| Arsenic | NMT 0.5 |
• Limit of Iron
65% Nitric acid:
Use ultratrace nitric acid, 65%–70% HNO3, ACS reagent grade.
Blank standard:
Prepared as directed in the Limit of Lead, Mercury, Chromium, Nickel, Cadmium, and Arsenic.
Standard stock solution 100 ppm:
Immediately before use, dilute an appropriate amount of iron standard4 with a solution of 65% Nitric acid (1 in 100) to prepare an acidic solution containing the equivalent of 100 µg/mL of iron.
Standard solutions:
Separately transfer 0.1 and 0.5 mL of Standard stock solution 100 ppm to 100-mL volumetric flasks, dilute with a solution of 65% Nitric acid (1 in 100) to volume, and mix. These solutions contain, respectively, 0.1 and 0.5 µg of iron/mL (Standard solution 0.1 ppm and Standard solution 0.5 ppm).
Sample solution:
Transfer 10.0 mL of the digestion solution from the test for Limit of Lead, Mercury, Chromium, Nickel, Cadmium, and Arsenic to a 100-mL volumetric flask, dilute with water to volume, and mix.
Blank solution:
Transfer 10.0 mL of the blank digestion solution from the test for Limit of Lead, Mercury, Chromium, Nickel, Cadmium, and Arsenic to a 100-mL volumetric flask, dilute with water to volume, and mix.
Instrumental conditions
(See Plasma Spectrochemistry  730
730 .)
.)
Mode:
Inductively coupled plasma–atomic emission spectrometer (ICP–AES)
Analytical wavelength:
238.040 and 239.562 nm with the settings optimized as directed by the manufacturer
Analysis
Samples:
Blank standard, Standard solutions, Sample solution, and Blank solution
Instrument performance must be verified to conform to the manufacturer’s specifications for resolution and sensitivity. Before analyzing samples, the instrument must pass a suitable performance check. Generate the calibration curve using the Blank standard, Standard solution 0.1 ppm, and Standard solution 0.5 ppm: the linear regression coefficient is NLT 0.999.
Aspirate the Blank solution and Sample solution, respectively, at least in duplicate, and report the average reading as the iron content of the sample. Determine the concentration CB, in µg/mL, of iron in the Blank solution, and also determine the concentration CS, in µg/mL, of iron in the Sample solution using the calibration curve.
Calculate the quantity, in µg/g, of iron in the portion of Chitosan taken:
Result = [(CS  CB)/W] × F × V
CB)/W] × F × V
| F | = | = dilution factor, 2 |
| V | = | = volume of the Sample solution, 100 mL |
| W | = | = weight of the Chitosan taken to prepare the Sample solution (g) |
Acceptance criteria:
NMT 10 ppm of iron
• Limit of Protein Content
Control solution A:
1.0 mg/mL of bovine serum albumin
Control solution B:
0.1 mg/mL of bovine serum albumin
Sample solution C:
Dissolve 100 mg of Chitosan in 4 mL of 100% formic acid, mix, and stir for about 48 h at room temperature using a magnetic stirrer. This solution contains 25 mg/mL of Chitosan in 100% formic acid.
Sample solutions D, E, F:
Dilute aliquots of the Sample solution C with water to obtain the following solutions with concentrations of 2.5 mg/mL of Chitosan in 10% formic acid, 0.5 mg/mL of Chitosan in 2% formic acid, and 0.25 mg/mL of Chitosan in 1% formic acid, respectively.
Molecular weight standard preparation G:
Use a commercially available preparation of apparent molecular weight protein standards of 10,000–190,000 Da dissolved in the loading buffer consisting of 50 mM of tris(hydroxymethyl)aminomethane hydrochloride (pH 6.8), 5 mM of ethylenediaminetetraacetic acid, 10 mM of dithiothreitol [Note—A 2%–5% solution of beta-mercaptoethanol can be used to replace dithiothreitol. ] , 1% (w/v) sodium dodecyl sulfate, and 10% (w/v) glycerol.5 [Note—A protein ladder containing the following molecular weight standards in 10, 15, 20, 25, 40, 50, 60, 85, 120, and 190 KDa or other appropriate combinations can be used. ]
Sample buffer:
Transfer 666 mg of tris(hydroxymethyl)aminomethane hydrochloride, 682 mg of tris(hydroxymethyl)aminomethane, 800 mg of lithium dodecyl sulfate, 6 mg of ethylenediaminetetraacetic acid, 4 g of glycerol, 0.75 mL of 1% solution of Coomassie blue G250, and 0.25 mL of 1% solution of phenol red to a 10-mL volumetric flask, add 8 mL of water to the flask, and mix. If necessary, adjust with hydrochloric acid or sodium hydroxide to pH of 7.2. Dilute with water to volume. [Note—Store the buffer at 4 . It is stable for 6 months when stored at 4
. It is stable for 6 months when stored at 4 . ] An equivalent commercially available buffer can also be used.6
. ] An equivalent commercially available buffer can also be used.6
Running buffer:
Prepare a solution containing 1 M of tris(hydroxymethyl)aminomethane, 1 M of 2-(4-morpholinyl) ethanesulfonic acid, 20.5 mM ethylenediaminetetraacetic acid, and 69.3 mM dodecyl sodium sulfate in water. If necessary, adjust with hydrochloric acid or sodium hydroxide to pH of 7.3. An equivalent commercially available appropriate SDS running buffer can be used.7
Fixing solution:
A solution containing 40% ethanol and 10% acetic acid
Sensitizing solution:
Transfer 10 mL of a solution mainly containing 10%–20% of 2-(4-morpholinyl) ethanesulfonic acid, 0.1%–1.0% of sodium hydroxide, and 7%–13% of N,N-dimethylformamide into a 100-mL volumetric flask, add 30 mL of alcohol, and dilute with water to volume. Alternatively, follow the instructions of a commercially available silver staining kit8 to prepare the Sensitizing solution, Staining solution, and Developing solution.
Staining solution:
Transfer 1.0 mL of a solution mainly containing 10%–30% of silver nitrate into a 100-mL volumetric flask, and dilute with water to volume.
Developing solution:
Transfer 10 mL of a solution mainly containing 10%–30% of sodium carbonate into a 100-mL volumetric flask, add 1 drop of a solution containing 30%–60% of formaldehyde, and dilute with water to volume.
Stopping solution:
A solution containing 10%–30% of ethylenediaminetetraacetic acid and 10%–30% of tris(hydroxymethyl)-aminomethane. A Stopping solution is commercially available and included in a silver staining kit.
Analysis:
Mix 75 µL each of Sample solutions C, D, E, and F with 25 µL of the Sample buffer, and incubate at 70 for 10 min. In a suitable device for polyacrylamide gel electrophoresis (see Electrophoresis
for 10 min. In a suitable device for polyacrylamide gel electrophoresis (see Electrophoresis  726
726 and Biotechnology-Derived Articles—Polyacrylamide Gel Electrophoresis
and Biotechnology-Derived Articles—Polyacrylamide Gel Electrophoresis  1056
1056 ) add appropriate volumes of the Running buffer in the upper and the lower buffer chambers. Attach a 4%–12% gradient Bis-Tris ready-made polyacrylamide gel sandwiched between two glass plates, such that the wells for sample application are exposed to the Running buffer in the upper buffer chamber.
) add appropriate volumes of the Running buffer in the upper and the lower buffer chambers. Attach a 4%–12% gradient Bis-Tris ready-made polyacrylamide gel sandwiched between two glass plates, such that the wells for sample application are exposed to the Running buffer in the upper buffer chamber.
Separately apply equal volumes (about 20 µL) of each of the treated Sample solutions C, D, E, and F, Control solution A, and Control solution B onto separate lanes; apply equal volumes (about 10 µL) of the Molecular weight standard preparation G to both sides of the gel. [Note—Do not apply any solution in the outside lanes. ] Connect the lower buffer chamber electrode to the positive terminal and the upper buffer chamber electrode to the negative terminal of a suitable power supply unit, and carry out the electrophoresis at a constant voltage of about 100 V for about 100 min.
Remove the gel from the gel assembly. [Note—Do not touch the gel with bare hands. Use gloves. ]
Place the gel in a clean staining tray of appropriate size. Rinse the gel briefly with water. Fix the gel in 100 mL of Fixing solution for 20 min with gentle rotation. [Note—The gel can be stored in the Fixing solution overnight. ]
Decant the Fixing solution, and wash the gel in 30% ethanol for 10 min. Decant the ethanol, and add 100 mL of Sensitizing solution to the washed gel in the staining container. Incubate the gel in the Sensitizing solution for about 10 min. [Note—All incubations should be performed on a rotary shaker rotating at a speed of 1 revolution/s at room temperature. ]
Decant the Sensitizing solution, and wash the gel in 100 mL of 30% ethanol for 10 min. Wash the gel in 100 mL of water for 10 min. Incubate the gel in 100 mL of Staining solution for 15 min. After staining is complete, decant the Staining solution, and wash the gel with 100 mL of water for 20–60 s. [Note—Washing the gel for more than 1 min will remove the silver ion from the gel resulting in decreased sensitivity. ]
Incubate the gel in 100 mL of Developing solution for 4–8 min until bands start to appear and the desired band intensity is reached.
Once the appropriate staining intensity is achieved, immediately add 10 mL of Stopping solution directly to the gel still immersed in the Developing solution. Gently agitate the gel for 10 min. The color changes from pink to colorless indicating that the development has stopped.
Decant the colorless solution, and wash the gel with 100 mL of water for 10 min.
Use a gel imaging system, ideally with a CCD camera, to record the results.
Acceptance criteria:
NMT 0.2% of protein
SPECIFIC TESTS
• Bacterial Endotoxins Test  85
85
Sample stock solution:
Transfer 0.5 g of Chitosan to a 50-mL volumetric flask, add LAL Reagent Water9 and 4.6 mL of 1 N hydrochloric acid, dilute with the LAL Reagent Water to volume, and mix well. Incubate this solution in a water bath at 40 for 48 h.
for 48 h.
Sample solution:
Dilute the Sample stock solution to 1:50 with LAL Reagent Water, including dilution 1:2 with a  -glucan blocker.10
-glucan blocker.10
Acceptance criteria:
The level of bacterial endotoxins is such that the requirement under the relevant dosage form monograph(s) in which Chitosan is used can be met. Where the label states that Chitosan must be subjected to further processing during the preparation of wound dressing dosage forms, the level of bacterial endotoxins is such that the requirement under the relevant dosage form monograph(s) in which Chitosan is used can be met.
• Microbial Enumeration Tests  61
61 and Tests for Specified Microorganisms
and Tests for Specified Microorganisms  62
62
Sample:
Prepare a solution (1 in 50).
Acceptance criteria:
The total aerobic microbial count does not exceed 103 cfu/g, and the total combined molds and yeasts count does not exceed 102 cfu/g. It meets the requirements of the tests for absence of Pseudomonas aeruginosa and Staphylococcus aureus.
• Apparent Average Molecular Weight and Molecular Weight Distribution
[Note—This test is applicable to Chitosan of an average molecular weight of NMT 1,000,000 Da. In the following test an apparent average molecular weight is determined. ]
Mobile phase:
Transfer 12.75 g of sodium nitrate to a 1000-mL volumetric flask containing 800 mL of water, add a suitable amount of formic acid, dilute with water to volume, mix well, and make a concentration of formic acid of 0.5 M. The Mobile phase contains 0.15 M sodium nitrate in 0.5 M aqueous formic acid.
System suitability solution:
1.0 mg/mL of ethylene glycol in Mobile phase
Standard solutions:
Prepare several sets of mixtures, containing ten polyethylene glycol (PEG) standards of different known molecular weight, which are used to cover the molecular weight range from about 200 to 1,100,000 g/mol.11 [Note—These standards could be mixtures of polyethylene glycols and polyethylene oxides. ] Prepare each set of PEG molecular weight standards to have a known concentration of about 1.0 mg/mL for each standard in Mobile phase. Allow the Standard solutions to stand at room temperature for at least 8 h. Do not filter before use.
Sample solution:
Prepare a solution containing 1.0 mg/mL of Chitosan in Mobile phase. Cap, and mix well. Allow the solution to stand at room temperature for at least 12 h. Pass the chitosan solution through a membrane filter of 0.45-µm pore size, discard an appropriate volume of the initial filtrate, and use the rest of the filtered solution for analysis.
Chromatographic system
Mode:
LC
Detector:
Refractive index
Detector temperature:
35
Columns:
7.5-mm × 30-cm analytical column, 10-µm packing L38; and two 7.5-mm × 30-cm analytical columns; 17-µm packing L38
Flow rate:
1.0 mL/min for System suitability solution; and 0.5 mL/min for Standard solutions and Sample solution
Injection size:
20 µL for System suitability solution; and 100 µL for Standard solutions and Sample solution
System suitability
Samples:
System suitability solution and Standard solution
Suitability requirements
Plate count:
NLT 80% of the value that is certified by the column manufacture for new columns, determining the plate count for the ethylene glycol peak, System suitability solution
Resolution:
NLT 1.7 between the PEG standards, Standard solution. [Note—The resolution between the PEG standard with a molecular weight of 1,000,000 Da and its adjacent PEG standard should be NLT 1.0 if 1.7 cannot be met. ]
Analysis
Samples:
Standards solutions and Sample solution
Determine elution peak maxima and corresponding retention volumes (elution volume), Vp, for the ten PEG standards, corresponding to the specified molecular weight, Mp, at the peak maximum of the standards.
Molecular weight calibration:
Analyze each polyethylene glycol standard, and use a suitable gel-permeation chromatography or size-exclusion chromatography (GPC/SEC) software, or an equivalent data handling system, to compute the data and calibration. Plot log Mp for each standard in the Standard solutions versus its retention volume, VP, in mL, at each standard peak maximum, and construct the best polynomial line fitting the ten points.
Data analysis for sample:
[Note—Based on the PEG molecular weight calibration curve, calculate the molecular weight and molecular weight distribution of Chitosan using the slice by slice method. ]
Analyze the Chitosan sample by identifying retention volumes Va and Vb corresponding to the beginning and end of the Chitosan chromatogram. The baseline between Va and Vb is assumed to be linear. [Note—Draw a straight line between Va and Vb. ] Data analysis is based on a suitable GPC/SEC computer software or a real-time data acquisition system with either off-line or on-line data processing that is able to provide a means of determining chromatographic peak heights or integrated area segments as prescribed intervals under the SEC chromatogram and handling and reporting the data. The following describes the data processes which can either be computed by the GPC/SEC software or by an equivalent data processing system. Upon acquisition, handle the data under the Chitosan elution peak in discrete segments Ai, integrated area slices, or as digitized chromatogram heights Hi by recording the vertical displacements between the chromatogram trace and the baseline at elution volume, Vi, over designated intervals. A minimum of 40 area segments or heights are required. Obtain the corresponding value of Mi for Chitosan based on its elution volume, Vi, from the molecular weight calibration curve obtained in Molecular weight calibration.
Calculate the number-, and weight-average molecular weights, Mn and Mw, in g/mol, respectively, using the following formulas.
If the elution volume internal DVi, for instance, V2  V1 = V3
V1 = V3  V2, etc, is constant; parameters Ai and Mi are the chromatographic peak slice area and Chitosan molecular weight associated with the elution volume, Vi; and N is the number of data points obtained from the chromatogram between Va and Vb (N
V2, etc, is constant; parameters Ai and Mi are the chromatographic peak slice area and Chitosan molecular weight associated with the elution volume, Vi; and N is the number of data points obtained from the chromatogram between Va and Vb (N  40). [Note—If N is sufficiently large, the use of area segments Ai or peak heights Hi will yield equivalent results. ]
40). [Note—If N is sufficiently large, the use of area segments Ai or peak heights Hi will yield equivalent results. ]
Calculate the molecular weight distribution or polydispersity for Chitosan using the following expression:
Result = Mw/Mn
Acceptance criteria:
The values of apparent weight-average molecular weight and polydispersity are NLT 85% and NMT 115% of their respective values stated on the label.
• Loss on Drying  731
731 :
Dry 1.0 g in an oven at 100
:
Dry 1.0 g in an oven at 100 –105
–105 for 7 h: it loses NMT 5.0% of its weight.
for 7 h: it loses NMT 5.0% of its weight.
ADDITIONAL REQUIREMENTS
• Packaging and Storage:
Preserve in light-resistant and well-closed containers in a dry place, and store at a temperature below 30 .
.
• Labeling:
Label it to indicate its apparent weight-average molecular weight, Mw, and polydispersity (Mw/Mn). Where Chitosan is intended for use in the manufacture of wound dressings, it is so labeled. Where Chitosan must be subjected to further processing during the preparation of wound dressings, it is so labeled. Label to indicate the natural source from which Chitosan is derived.
1
A suitable yttrium ICP standard is available from LGC Promochem (www.lgcpromochem.com) or from Merck KGaA, Frankfurter Str. 250, 64293 Darmstadt, Germany.
2
A suitable lutetium ICP standard is available from LGC Promochem (www.lgcpromochem.com) or from Merck KGaA, Frankfurter Str. 250, 64293 Darmstadt, Germany.
3
Suitable multi-element ICP standard solutions are available from LGC Promochem (www.lgcpromochem.com) or from Merck KGaA, Frankfurter Str. 250, 64293 Darmstadt, Germany.
4
Suitable iron standards are available from LGC Promochem (www.lgcpromochem.com) (Single element standard for ICP, Iron 10,000 µg/mL dilute nitric acid) or from Merck KGaA, Frankfurter Str. 250, 64293 Darmstadt, Germany (iron ICP standard, 10000 mg/L in 10% nitric acid).
5
A suitable molecular weight standard preparation is available as BenchMark Prestained Protein Ladder from Invitrogen, product number: 10748010.
6
A suitable sample buffer is available as 4X NuPAGE LDS sample buffer from Invitrogen, product number: NP0007.
7
A suitable running buffer is available as NuPAGE MES SDS Running Buffer from Invitrogen, product number: NP0002.
8
A suitable silver staining kit is available as NuPAGE Silver Staining Kit from Invitrogen, product number: LC6070.
9
Use Sterile Water for Injection or other water that shows no reaction with the specific LAL Reagent with which it is to be used, at the limit of sensitivity of such reagent.
10
Available from Cambrex Europe s.p.r.l., Verviers, Belgium; Charles River, www.criver.com; or all the major LAL manufacturers.
11
Suitable polyethylene glycol molecular weight standards are available as ReadyCal kits from Polymer Standards Service (PSS), www.polymer.de.
Auxiliary Information—
Please check for your question in the FAQs before contacting USP.
| Topic/Question | Contact | Expert Committee |
|---|---|---|
| Monograph | Robert H. Lafaver, M.S.
Scientific Liaison 1-301-816-8335 |
(EXC2010) Monographs - Excipients |
| Radhakrishna S Tirumalai, Ph.D.
Principal Scientific Liaison 1-301-816-8339 |
(GCM2010) General Chapters - Microbiology | |
| Radhakrishna S Tirumalai, Ph.D.
Principal Scientific Liaison 1-301-816-8339 |
(GCM2010) General Chapters - Microbiology | |
| Radhakrishna S Tirumalai, Ph.D.
Principal Scientific Liaison 1-301-816-8339 |
(GCM2010) General Chapters - Microbiology | |
| Reference Standards | RS Technical Services 1-301-816-8129 rstech@usp.org |
USP35–NF30 Page 1754
Pharmacopeial Forum: Volume No. 35(1) Page 115