Delete the following:
Scope
Polyacrylamide gel electrophoresis is used for the qualitative characterization of proteins in biological preparations, for control of purity, and for quantitative determinations.
Purpose
Analytical gel electrophoresis is an appropriate method with which to identify and to assess the homogeneity of proteins in pharmaceutical preparations. The method is routinely used for the estimation of protein subunit molecular masses and for determination of the subunit compositions of purified proteins. Ready-to-use gels and reagents are commercially available and can be used instead of those described in this text, provided that they give equivalent results and that they meet the validity requirements given in Validation of the Test (below). 2S (USP38)
2S (USP38)
Delete the following:
The sieving properties of polyacrylamide gels are established by the three-dimensional network of fibers and pores that is formed as the bifunctional bisacrylamide cross-links adjacent polyacrylamide chains. Polymerization usually is catalyzed by a free radical–generating system composed of ammonium persulfate and N,N,N¢,N¢-tetramethylethylenediamine (TEMED).
As the acrylamide concentration of a gel increases, its effective pore size decreases. The effective pore size of a gel is operationally defined by its sieving properties, that is, by the resistance it imparts to the migration of macromolecules. There are limits on the acrylamide concentrations that can be used. As the pore size of a gel decreases, the migration rate of a protein through the gel decreases. By adjusting the pore size of a gel through manipulating the acrylamide concentration, analysts can optimize the resolution of the method for a given protein product. Thus, a given gel is physically characterized by its respective composition of acrylamide and bisacrylamide.
In addition to the composition of the gel, the state of the protein is an important component of electrophoretic mobility. In the case of proteins, the electrophoretic mobility depends on the pK value of the charged groups and the size of the molecule. It is influenced by the type, the concentration, and the pH of the buffer; by the temperature and the field strength; and by the nature of the support material. 2S (USP38)
2S (USP38)
Delete the following:
The method cited as an example is limited to the analysis of monomeric polypeptides with a mass range of 14,000–100,000 Da. It is possible to extend this mass range by various techniques (e.g., gradient gels and particular buffer system). For instance, tricine–sodium dodecyl sulfate (SDS) gels, using tricine instead of glycine (in the method described here) as the trailing ion, can separate very small proteins and peptides under 10,000–15,000 Da.
Denaturing polyacrylamide gel electrophoresis using glycine sodium dodecyl sulfate (SDS-PAGE) is the most common mode of electrophoresis used in assessing the pharmaceutical quality of protein products and is the focus of the example method. Typically, analytical electrophoresis of proteins is carried out in polyacrylamide gels under conditions that ensure dissociation of the proteins into their individual polypeptide subunits and that minimize aggregation. Most commonly, the strongly anionic detergent sodium dodecyl sulfate (SDS) is used in combination with heat to dissociate the proteins before they are loaded on the gel. The denatured polypeptides bind to SDS, become negatively charged, and exhibit a consistent charge-to-mass ratio regardless of protein type. Because the amount of SDS bound is almost always proportional to the molecular mass of the polypeptide and is independent of its sequence, SDS–polypeptide complexes migrate through polyacrylamide gels with mobilities dependent on the size of the polypeptide.
The electrophoretic mobilities of the resultant detergent–polypeptide complexes all assume the same functional relationship to their molecular masses. SDS complexes migrate toward the anode in a predictable manner; low-molecular-mass complexes migrate faster than larger ones. The molecular mass of a protein therefore can be estimated from its relative mobility in calibrated SDS-PAGE, and the intensity of a single band relative to other undesired bands in such a gel can be a measure of purity.
Modifications to the polypeptide backbone, such as N- or O-linked glycosylation, can change the apparent molecular mass of a protein, because SDS does not bind to a carbohydrate moiety in a manner similar to that of a polypeptide; therefore, a consistent charge-to-mass ratio is not maintained.
Depending on the extent of glycosylation and other posttranslational modifications, the apparent molecular mass of proteins may not be a true reflection of the mass of the polypeptide chain.
Reducing Conditions
Polypeptide subunits and three-dimensional structure often are maintained in proteins by the presence of disulfide bonds. A goal of SDS-PAGE analysis under reducing conditions is to disrupt this structure by reducing disulfide bonds. Complete denaturation and dissociation of proteins by treatment with 2-mercaptoethanol (2-ME) or dithiothreitol (DTT) results in unfolding of the polypeptide backbone and subsequent complexation with SDS. Using these conditions, analysts can reasonably calculate the molecular mass of the polypeptide by linear regression (or, more closely, by nonlinear regression) in the presence of suitable molecular mass standards.
Nonreducing Conditions
For some analyses, complete dissociation of the protein into subunit peptides is not desirable. In the absence of treatment with reducing agents such as 2-ME or DTT, disulfide covalent bonds remain intact, preserving the oligomeric form of the protein. Oligomeric SDS–protein complexes migrate more slowly than their SDS–polypeptide subunits. In addition, nonreduced proteins may not be completely saturated with SDS and hence may not bind the detergent in a constant mass ratio. Moreover, intrachain disulfide bonds constrain the molecular shape, usually in such a way that reduces the Stokes radius of the molecule, thereby reducing the apparent molecular mass, Mr. This makes molecular mass determinations of these molecules by SDS-PAGE less straightforward than analyses of fully denatured polypeptides because both standards and unknown proteins must be present in similar configurations for valid comparisons. 2S (USP38)
2S (USP38)
Delete the following:
The most popular electrophoretic method for the characterization of complex mixtures of proteins uses a discontinuous buffer system involving two contiguous but distinct gels: a resolving or separating (lower) gel and a stacking (upper) gel. The two gels are cast with different porosities, pH, and ionic strengths. In addition, different mobile ions are used in the gel and electrode buffers. The buffer discontinuity acts to concentrate large-volume samples in the stacking gel, resulting in improved resolution. When power is applied, a voltage drop develops across the sample solution and drives the proteins into the stacking gel. Glycinate ions from the electrode buffer follow the proteins into the stacking gel. A moving boundary region is rapidly formed, with the highly mobile chloride ions in the front and the relatively slow glycinate ions in the rear. A localized high-voltage gradient forms between the leading and trailing ion fronts, causing the SDS–protein complexes to form into a thin zone (stack) and to migrate between the chloride and glycinate phases. Within broad limits, regardless of the height of the applied sample, all SDS–proteins condense into a very narrow region and enter the resolving gel as a well-defined, thin zone of high protein density. The large-pore stacking gel does not retard the migration of most proteins and serves mainly as an anticonvective medium. At the interface of the stacking and resolving gels, the proteins experience a sharp increase in retardation due to the restrictive pore size of the resolving gel and the buffer discontinuity, which also contributes to unstacking of the proteins. Once in the resolving gel, proteins continue to be slowed by the sieving of the matrix. The glycinate ions overtake the proteins, which then move in a space of uniform pH formed by the tris(hydroxymethyl)aminomethane (Tris) and glycine. Molecular sieving causes the SDS–polypeptide complexes to separate on the basis of their molecular masses. 2S (USP38)
2S (USP38)
Delete the following:
This section describes the preparation of gels using particular instrumentation. This does not apply to precast gels. For precast gels or any other commercially available equipment, the manufacturer's instructions should be used for guidance.
The use of commercial reagents that have been purified in solution is recommended. When this is not the case and when the purity of the reagents used is not sufficient, a pretreatment is applied. For instance, any solution sufficiently impure to require filtration must also be deionized with a mixed-bed (anion–cation exchange) resin to remove acrylic acid and other charged degradation products. Unopened, gas-sparged (with argon or nitrogen) acrylamide–bisacrylamide solutions and persulfate solid that is kept dry in a desiccator or a sealed bottle containing silica gel are stable for long periods. Fresh ammonium persulfate solutions are prepared daily.
Gel Stock Solutions
30% acrylamide–bisacrylamide solution
Prepare a solution containing 290 g of acrylamide and 10 g of methylenebisacrylamide per liter of water. Filter.
ammonium persulfate solution
Prepare a small quantity of solution having a concentration of 100 g/L of ammonium persulfate. [Note—Ammonium persulfate provides the free radicals that drive polymerization of acrylamide and bisacrylamide. Because ammonium persulfate decomposes rapidly, fresh solutions must be prepared daily.
temed
Use an electrophoresis-grade reagent.
sds solution
This is a 100 g/L solution of electrophoresis-grade SDS.
1.5 m buffer solution
Dissolve 90.8 g of Tris in 400 mL of water. Adjust the pH to 8.8 with hydrochloric acid, and dilute to 500.0 mL with water.
1 m buffer solution
Dissolve 60.6 g of Tris in 400 mL of water. Adjust the pH to 6.8 with hydrochloric acid, and dilute to 500.0 mL with water.
Assembling the Gel-Molding Cassette
Clean the two glass plates (size, e.g., 10 cm × 8 cm), the polytetrafluoroethylene comb, the two spacers, and the silicone rubber tubing (e.g., 0.6 mm diameter × 35 cm length) with mild detergent; rinse extensively with water, followed by dehydrated alcohol; and allow the plates to dry at room temperature. Note that drying with a towel or a tissue may introduce stainable contamination, whereas using air drying prevents this risk. Lubricate the spacers and the tubing with nonsilicone grease. Apply the spacers along each of the two short sides of the glass plate 2 mm away from the edges and 2 mm away from the long side corresponding to the bottom of the gel. Begin to lay the tubing on the glass plate by using one spacer as a guide. Carefully twist the tubing at the bottom of the spacer, and follow the long side of the glass plate. While holding the tubing with one finger along the long side, twist the tubing again and lay it on the second short side of the glass plate, using the spacer as a guide. Place the second glass plate in perfect alignment, and hold the mold together by hand pressure.
Apply two clamps on each of the two short sides of the mold. Carefully apply four clamps on the longer side of the gel mold, thus forming the bottom of the gel mold. Verify that the tubing is running along the edge of the glass plates and has not been extruded while the clamps were placed. The gel mold is now ready for pouring the gel.
Preparation of the Gel
In a discontinuous buffer SDS polyacrylamide gel, it is recommended to pour the resolving gel, let the gel set, and then pour the stacking gel, because the composition of the two gels in acrylamide–bisacrylamide, buffer, and pH are different.
preparation of the resolving gel
In a conical flask, prepare the appropriate volume of solution containing the desired concentration of acrylamide for the resolving gel, using the values given in Table 1. Mix the components in the order shown. Where appropriate, before adding the Ammonium Persulfate Solution and the TEMED, filter the solution if necessary under vacuum through a cellulose acetate membrane (pore diameter: 0.45 µm). Keep the solution under vacuum, while swirling the filtration unit, until no more bubbles are formed in the solution. Add appropriate amounts of Ammonium Persulfate Solution and TEMED, as indicated in Table 1, swirl, and pour immediately into the gap between the two glass plates of the mold. Leave sufficient space for the stacking gel (the length of the teeth of the comb plus 1 cm). Using a tapered glass pipet, carefully overlay the solution with water-saturated isobutanol. Leave the gel in a vertical position at room temperature to allow polymerization.
Table 1. Preparation of the Resolving Gel
| |
Component Volume (mL) per Gel Mold Volume Below |
|||||||
|---|---|---|---|---|---|---|---|---|
| Solution component |
5 mL |
10 mL |
15 mL |
20 mL |
25 mL |
30 mL |
40 mL |
50 mL |
| 6% Acrylamide |
|
|
|
|
|
|
|
|
| 2.6 |
5.3 |
7.9 |
10.6 |
13.2 |
15.9 |
21.2 |
26.5 |
|
Solution |
1.0 |
2.0 |
3.0 |
4.0 |
5.0 |
6.0 |
8.0 |
10.0 |
| 1.3 |
2.5 |
3.8 |
5.0 |
6.3 |
7.5 |
10.0 |
12.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.004 |
0.008 |
0.012 |
0.016 |
0.02 |
0.024 |
0.032 |
0.04 |
|
| |
|
|
|
|
|
|
|
|
| 8% Acrylamide |
|
|
|
|
|
|
|
|
| 2.3 |
4.6 |
6.9 |
9.3 |
11.5 |
13.9 |
18.5 |
23.2 |
|
Solution |
1.3 |
2.7 |
4.0 |
5.3 |
6.7 |
8.0 |
10.7 |
13.3 |
| 1.3 |
2.5 |
3.8 |
5.0 |
6.3 |
7.5 |
10.0 |
12.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.003 |
0.006 |
0.009 |
0.012 |
0.015 |
0.018 |
0.024 |
0.03 |
|
| |
|
|
|
|
|
|
|
|
| 10% Acrylamide |
|
|
|
|
|
|
|
|
| 1.9 |
4.0 |
5.9 |
7.9 |
9.9 |
11.9 |
15.9 |
19.8 |
|
Solution |
1.7 |
3.3 |
5.0 |
6.7 |
8.3 |
10.0 |
13.3 |
16.7 |
| 1.3 |
2.5 |
3.8 |
5.0 |
6.3 |
7.5 |
10.0 |
12.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.002 |
0.004 |
0.006 |
0.008 |
0.01 |
0.012 |
0.016 |
0.02 |
|
| |
|
|
|
|
|
|
|
|
| 12% Acrylamide |
|
|
|
|
|
|
|
|
| 1.6 |
3.3 |
4.9 |
6.6 |
8.2 |
9.9 |
13.2 |
16.5 |
|
Solution |
2.0 |
4.0 |
6.0 |
8.0 |
10.0 |
12.0 |
16.0 |
20.0 |
| 1.3 |
2.5 |
3.8 |
5.0 |
6.3 |
7.5 |
10.0 |
12.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.002 |
0.004 |
0.006 |
0.008 |
0.01 |
0.012 |
0.016 |
0.02 |
|
| |
|
|
|
|
|
|
|
|
| 14% Acrylamide |
|
|
|
|
|
|
|
|
| 1.4 |
2.7 |
3.9 |
5.3 |
6.6 |
8.0 |
10.6 |
13.8 |
|
Solution |
2.3 |
4.6 |
7.0 |
9.3 |
11.6 |
13.9 |
18.6 |
23.2 |
| 1.2 |
2.5 |
3.6 |
5.0 |
6.3 |
7.5 |
10.0 |
12.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.002 |
0.004 |
0.006 |
0.008 |
0.01 |
0.012 |
0.016 |
0.02 |
|
| |
|
|
|
|
|
|
|
|
| 15% Acrylamide |
|
|
|
|
|
|
|
|
| 1.1 |
2.3 |
3.4 |
4.6 |
5.7 |
6.9 |
9.2 |
11.5 |
|
Solution |
2.5 |
5.0 |
7.5 |
10.0 |
12.5 |
15.0 |
20.0 |
25.0 |
| 1.3 |
2.5 |
3.8 |
5.0 |
6.3 |
7.5 |
10.0 |
12.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.002 |
0.004 |
0.006 |
0.008 |
0.01 |
0.012 |
0.016 |
0.02 |
|
preparation of the stacking gel
After polymerization is complete (about 30 min), pour off the isobutanol, and wash the top of the gel several times with water to remove the isobutanol overlay and any unpolymerized acrylamide. Drain as much fluid as possible from the top of the gel, and then remove any remaining water with the edge of a paper towel.
In a conical flask, prepare the appropriate volume of solution containing the desired concentration of acrylamide, using the values given in Table 2. Mix the components in the order shown. Where appropriate, before adding the Ammonium Persulfate Solution and the TEMED, filter the solution if necessary under vacuum through a cellulose acetate membrane (pore diameter: 0.45 µm). Keep the solution under vacuum, while swirling the filtration unit, until no more bubbles are formed in the solution. Add appropriate amounts of Ammonium Persulfate Solution and TEMED, as indicated in Table 2. Swirl, and pour immediately into the gap between the two glass plates of the mold directly onto the surface of the polymerized resolving gel. Immediately insert a clean polytetrafluoroethylene comb into the stacking gel solution, being careful to avoid trapping air bubbles. Add more stacking gel solution to fill the spaces of the comb completely. Leave the gel in a vertical position, and allow it to polymerize at room temperature.
Table 2. Preparation of the Stacking Gel
| |
Component Volume (mL) per Gel Mold Volume Below |
|||||||
|---|---|---|---|---|---|---|---|---|
| Solution component |
1 mL |
2 mL |
3 mL |
4 mL |
5 mL |
6 mL |
8 mL |
10 mL |
| Water |
0.68 |
1.4 |
2.1 |
2.7 |
3.4 |
4.1 |
5.5 |
6.8 |
| 30% Acrylamide–Bisacrylamide
Solution |
0.17 |
0.33 |
0.5 |
0.67 |
0.83 |
1.0 |
1.3 |
1.7 |
| 1.0 M Buffer Solution
|
0.13 |
0.25 |
0.38 |
0.5 |
0.63 |
0.75 |
1.0 |
1.25 |
| SDS Solution
|
0.01 |
0.02 |
0.03 |
0.04 |
0.05 |
0.06 |
0.08 |
0.1 |
| Ammonium Persulfate Solution
|
0.01 |
0.02 |
0.03 |
0.04 |
0.05 |
0.06 |
0.08 |
0.1 |
| TEMED
|
0.001 |
0.002 |
0.003 |
0.004 |
0.005 |
0.006 |
0.008 |
0.01 |
Preparation of the Sample
Unless otherwise specified in the specific monograph, the samples can be prepared as follows:
sds-page sample buffer (concentrated)
Dissolve 1.89 g of Tris, 5.0 g of sodium lauryl sulfate, and 50 mg of bromophenol blue in water. Add 25.0 mL of glycerol, and dilute to 100 mL with water. Adjust the pH to 6.8 with hydrochloric acid, and dilute to 125 mL with water.
sds-page sample buffer for reducing conditions (concentrated)
Dissolve 3.78 g of Tris, 10.0 g of SDS, and 100 mg of bromophenol blue in water. Add 50.0 mL of glycerol, and dilute to 200 mL with water. Add 25.0 mL of 2-ME. Adjust to pH 6.8 with hydrochloric acid, and dilute to 250.0 mL with water. Alternatively, DTT can be used as reducing agent instead of 2-ME. In this case prepare the sample buffer as follows: Dissolve 3.78 g of Tris, 10.0 g of SDS, and 100 mg of bromophenol blue in water. Add 50.0 mL of glycerol, and dilute to 200 mL with water. Adjust to pH 6.8 with hydrochloric acid, and dilute to 250.0 mL with water. Immediately before use, add DTT to a final concentration of 100 mM.
sds-page running buffer
Dissolve 151.4 g of Tris, 721.0 g of glycine, and 50.0 g of sodium lauryl sulfate in water, and dilute to 5000 mL with the same solvent. Immediately before use, dilute to 10 times its volume with water, and mix. Measure the pH of the diluted solution. The pH is between 8.1 and 8.8.
sample buffer (nonreducing conditions)
Mix equal volumes of water and SDS-PAGE Sample Buffer (Concentrated).
sample buffer (reducing conditions)
Mix equal volumes of water and SDS-PAGE Sample Buffer for Reducing Conditions (Concentrated) containing 2-ME (or DTT) as the reducing agent.
Dilute the preparation to be examined and the reference solutions with sample buffer to obtain the concentration prescribed in the monograph (depending on the protein and staining method, this concentration can vary).
Sample treatment: Boil for 5 min or use a block heater, and then chill. (Note that temperature and time may vary in the monograph because protein cleavage may occur during the heat treatment.)
Mounting the Gel in the Electrophoresis Apparatus and Electrophoretic Separation
After polymerization is complete (about 30 min), remove the polytetrafluoroethylene comb carefully. Rinse the wells immediately with water or with theSDS-PAGE Running Buffer to remove any unpolymerized acrylamide. If necessary, straighten the teeth of the stacking gel with a blunt hypodermic needle attached to a syringe. Remove the clamps on one short side, carefully pull out the tubing, and replace the clamps. Proceed similarly on the other short side. Remove the tubing from the bottom part of the gel. Mount the gel in the electrophoresis apparatus. Add the electrophoresis buffers to the top and bottom reservoirs. Remove any bubbles that become trapped at the bottom of the gel between the glass plates. This is best done with a bent hypodermic needle attached to a syringe. Never prerun the gel before loading the samples, because this will destroy the discontinuity of the buffer systems. Before loading the sample, carefully rinse each well with SDS-PAGE Running Buffer. Prepare the test and reference solutions in the recommended sample buffer, and treat as specified in the individual monograph. Apply the appropriate volume of each solution to the stacking gel wells.
Start the electrophoresis using the conditions recommended by the equipment manufacturer. Manufacturers of SDS-PAGE equipment may provide gels of different surface area and thickness, and electrophoresis running time and current or voltage may vary in order to achieve optimal separation. Check that the dye front is moving into the resolving gel. When the dye is near the bottom of the gel stop the electrophoresis. Remove the gel assembly from the apparatus, and carefully separate the glass plates. Remove the spacers, cut off and discard the stacking gel, and immediately proceed with staining. 2S (USP38)
2S (USP38)
Delete the following:
Gradient gels (resolving gels) are prepared with an increasing concentration of acrylamide from the top to the bottom. Preparation of gradient gels requires a gradient-forming apparatus. Ready-to-use gradient gels are commercially available with specific recommended protocols.
Gradient gels offer some advantages over fixed-concentration gels. Some proteins that co-migrate on fixed-concentration gels can be resolved within gradient gels. During electrophoresis the proteins migrate until the pore size stops further progress, and therefore a stacking effect occurs, resulting in sharper bands. According to Table 3, gradient gels also allow separation of a wider range of protein molecular masses than do single, fixed-concentration gels.
Table 3 gives suggested compositions of the linear gradient, relating the range of acrylamide concentrations to the appropriate protein molecular ranges. Note that other gradient shapes (e.g., concave) can be prepared for specific applications.
Table 3. Acrylamide Gradient Percentages Recommended for
Expected Protein Molecular Weights
Expected Protein Molecular Weights
| Acrylamide (%) |
Protein Range (kDa) |
|---|---|
| 5–15 |
20–250 |
| 5–20 |
10–200 |
| 10–20 |
10–150 |
| 8–20 |
8–150 |
Gradient gels also are used for molecular mass determination and protein purity determination. 2S (USP38)
2S (USP38)
Delete the following:
Coomassie and silver staining are the most common protein staining methods and are described in more detail below. Several other commercial stains, detection methods, and commercial kits are available. For example, fluorescent stains are visualized using a fluorescent imager and often provide a linear response over a wide range of protein concentrations—often several orders of magnitude, depending on the protein.
Coomassie staining has a protein detection level of approximately 1–10 µg of protein per band. Silver staining is the most sensitive method for staining proteins in gels, and a band containing 10–100 ng can be detected. These figures are considered robust in the context of these gels. Improved sensitivity of one or two orders of magnitude has been reported in the literature.
Coomassie staining responds in a more linear manner than silver staining, but the response and range depend on the protein and development time. Both Coomassie and silver staining can be less reproducible if staining is stopped in a subjective manner, i.e., when the analyst deems the staining satisfactory. Wide dynamic ranges of reference proteins are important to use because they help assess the intra-experimental sensitivity and linearity. All gel-staining steps are done while wearing gloves, at room temperature, with gentle shaking (e.g., on an orbital shaker platform), and using any convenient container.
Staining Reagents
destaining solution
Prepare a mixture of 1 volume of glacial acetic acid, 4 volumes of methanol, and 5 volumes of water.
coomassie staining solution
Prepare a 1.25 g/L solution of acid blue 83 in Destaining Solution. Filter.
fixing solution
To 250 mL of methanol, add 0.27 mL of formaldehyde, and dilute to 500.0 mL with water.
silver nitrate reagent
To a mixture of 3 mL of concentrated ammonia and 40 mL of 1 M sodium hydroxide, add 8 mL of a 200 g/L solution of silver nitrate, dropwise, with stirring. Dilute to 200 mL with water.
developer solution
Dilute 2.5 mL of a 20 g/L solution of citric acid and 0.27 mL of formaldehyde to 500.0 mL with water.
blocking solution
A 10% (v/v) solution of acetic acid.
Coomassie Staining
Immerse the gel in a large excess of Coomassie Staining Solution, and allow to stand for at least 1 h. Remove the staining solution.
Destain the gel with a large excess of Destaining Solution. Change the Destaining Solution several times until the stained protein bands are clearly distinguishable on a clear background. The more thoroughly the gel is destained, the smaller is the amount of protein that can be detected by the method. More rapid destaining can be achieved by including a few grams of anion-exchange resin or a small sponge in the Destaining Solution. [Note—The acid–alcohol solutions used in this procedure do not completely fix proteins in the gel. This can lead to losses of some low-molecular-mass proteins during the staining and destaining of thin gels. Permanent fixation is obtainable by allowing the gel to stand in a mixture of 1 volume of trichloroacetic acid, 4 volumes of methanol, and 5 volumes of water for 1 h before it is immersed in the Coomassie Staining Solution.
Silver Staining
Immerse the gel in a large excess of Fixing Solution, and allow it to stand for 1 h. Remove the Fixing Solution, add fresh Fixing Solution, and incubate for at least 1 h or overnight, if convenient. Discard the Fixing Solution, and wash the gel in a large excess of water for 1 h. Soak the gel for 15 min in a 1% (v/v) solution of glutaraldehyde. Wash the gel twice for 15 min in a large excess of water. Soak the gel in fresh Silver Nitrate Reagent for 15 min, in darkness. Wash the gel three times for 5 min in a large excess of water. Immerse the gel for about 1 min in Developer Solution until satisfactory staining has been obtained. Stop the development by incubation in the Blocking Solution for 15 min. Rinse the gel with water. 2S (USP38)
2S (USP38)
Delete the following:
Gels are photographed or scanned while they are still wet or after an appropriate drying procedure. Currently, gel-scanning systems with data analysis software are commercially available to photograph and analyze the wet gel immediately.
Drying of stained SDS polyacrylamide gels produces permanent documentation. This method, however, frequently results in gel cracking during drying between cellulose films, rendering the gel unsuitable for any kind of densitometry analyses later.
Depending on the staining method used, gels are treated in a slightly different way. For Coomassie staining, after the destaining step, allow the gel to stand in a 100 g/L solution of glycerol for at least 2 h (overnight incubation is possible). For silver staining, add to the final rinsing a step of 5 min in a 20 g/L solution of glycerol.
Immerse two sheets of porous cellulose film in water, and incubate for 5–10 min.
Place one of the sheets on a drying frame. Carefully lift the gel, and place it on the cellulose film. Remove any trapped air bubbles, and pour a few mL of water around the edges of the gel. Place the second sheet on top, and remove any trapped air bubbles. Complete the assembly of the drying frame. Place in an oven or leave at room temperature until dry. 2S (USP38)
2S (USP38)
Delete the following:
Molecular masses of proteins are determined by comparison of their mobilities with those of several marker proteins of known molecular weight. Mixtures of prestained and unstained proteins with precisely known molecular masses blended for uniform staining are available for calibrating gels. They are available in various molecular mass ranges. Concentrated stock solutions of proteins of known molecular mass are diluted in the appropriate sample buffer and are loaded on the same gel as the protein sample to be studied.
Immediately after the gel has been run, mark the position of the bromophenol blue tracking dye to identify the leading edge of the electrophoretic ion front. This can be done by cutting notches in the edges of the gel or by inserting a needle soaked in India ink into the gel at the dye front. After staining, measure the migration distances of each protein band (markers and unknowns) from the top of the resolving gel. Divide the migration distance of each protein by the distance traveled by the tracking dye. The normalized migration distances are referred to as the relative mobilities of the proteins (relative to the dye front), or RF. Construct a plot of the logarithm of the relative molecular masses (Mr) of the protein standards as a function of the RF values. Unknown molecular masses can be estimated by linear regression analysis (more accurately, by nonlinear regression analysis) or interpolation from the curves of log Mr against RF if the values obtained for the unknown samples are positioned along the approximately linear part of the graph. 2S (USP38)
2S (USP38)
Delete the following:
The test is not valid unless the proteins of the molecular mass marker are distributed along 80% of the length of the gel and over the required separation range covering the relevant protein bands (e.g., the product and the dimer or the product and its related impurities). The separation obtained for the expected proteins must show a linear relationship between the logarithm of the molecular mass and the RF. If the plot has a sigmoidal shape, then only data from the linear region of the curve can be used in the calculations. Additional validation requirements with respect to the test sample may be specified in individual monographs.
Sensitivity also must be validated. A reference protein control corresponding to the desired concentration limit that is run in parallel with the test samples can serve as a system suitability check of the experiment. 2S (USP38)
2S (USP38)
Delete the following:
When impurities are quantitated by normalization to the main band using an integrating densitometer or image analysis, the responses must be validated for linearity. Note that depending on the detection method and protein, as described in the introduction of the section Detection of Proteins in Gels, the linear range can vary but can be assessed within each run by using one or more control samples containing an appropriate range of protein concentrations.
When the impurity limit is specified in the individual monograph, analysts should prepare a reference solution corresponding to that level of impurity by diluting the test solution. For example, when the limit is 5%, a reference solution would be a 1:20 dilution of the test solution. No impurity (any band other than the main band) in the electropherogram obtained with the test solution may be more intense than the main band obtained with the reference solution.
Under validated conditions, impurities can be quantified by normalization to the main band, using an integrating densitometer, or by image analysis. 2S (USP38)
2S (USP38)
Add the following:
Scope
Polyacrylamide gel electrophoresis is used for the qualitative characterization of proteins in biological preparations, for control of purity, and for quantitative determinations.
Purpose
Analytical gel electrophoresis is an appropriate method with which to identify and to assess the homogeneity of proteins in pharmaceutical preparations. The method is routinely used for the estimation of protein subunit molecular masses and for determination of the subunit compositions of purified proteins. Ready-to-use gels and reagents are commercially available and can be used instead of those described in this text, provided that they give equivalent results and that they meet the validity requirements given in Validation of the Test (below).
As the acrylamide concentration of a gel increases, its effective pore size decreases. The effective pore size of a gel is operationally defined by its sieving properties, that is, by the resistance it imparts to the migration of macromolecules. There are limits on the acrylamide concentrations that can be used. At high acrylamide concentrations, gels break much more easily and are difficult to handle. As the pore size of a gel decreases, the migration rate of a protein through the gel decreases. By adjusting the pore size of a gel through manipulating the acrylamide concentration, analysts can optimize the resolution of the method for a given protein product. Thus, a given gel is physically characterized by its respective composition of acrylamide and bisacrylamide.
In addition to the composition of the gel, the state of the protein is an important component of electrophoretic mobility. In the case of proteins, the electrophoretic mobility depends on the pK value of the charged groups and the size of the molecule. It is influenced by the type, the concentration, and the pH of the buffer; by the temperature and the field strength; and by the nature of the support material.
Denaturing polyacrylamide gel electrophoresis using glycine SDS (SDS-PAGE) is the most common mode of electrophoresis used in assessing the pharmaceutical quality of protein products and is the focus of the example method. Typically, analytical electrophoresis of proteins is carried out in polyacrylamide gels under conditions that ensure dissociation of the proteins into their individual polypeptide subunits and that minimize aggregation. Most commonly, the strongly anionic detergent SDS is used in combination with heat to dissociate the proteins before they are loaded on the gel. The denatured polypeptides bind to SDS, become negatively charged, and exhibit a consistent charge-to-mass ratio regardless of protein type. Because the amount of SDS bound is almost always proportional to the molecular mass of the polypeptide and is independent of its sequence, SDS–polypeptide complexes migrate through polyacrylamide gels with mobilities dependent on the size of the polypeptide.
The electrophoretic mobilities of the resultant detergent–polypeptide complexes all assume the same functional relationship to their molecular masses. SDS complexes migrate toward the anode in a predictable manner; low-molecular-mass complexes migrate faster than larger ones. The molecular mass of a protein therefore can be estimated from its relative mobility in calibrated SDS-PAGE, and the intensity of a single band relative to other undesired bands in such a gel can be a measure of purity.
Modifications to the polypeptide backbone, such as N- or O-linked glycosylation, can change the apparent molecular mass of a protein, because SDS does not bind to a carbohydrate moiety in a manner similar to that of a polypeptide; therefore, a consistent charge-to-mass ratio is not maintained.
Depending on the extent of glycosylation and other posttranslational modifications, the apparent molecular mass of proteins may not be a true reflection of the mass of the polypeptide chain.
Reducing Conditions
Polypeptide subunits and three-dimensional structure often are maintained in proteins by the presence of disulfide bonds. A goal of SDS-PAGE analysis under reducing conditions is to disrupt this structure by reducing disulfide bonds. Complete denaturation and dissociation of proteins by treatment with 2-mercaptoethanol (2-ME) or dithiothreitol (DTT) results in unfolding of the polypeptide backbone and subsequent complexation with SDS. Using these conditions, analysts can reasonably calculate the molecular mass of the polypeptide by linear regression (or, more closely, by nonlinear regression) in the presence of suitable molecular mass standards.
Nonreducing Conditions
For some analyses, complete dissociation of the protein into subunit peptides is not desirable. In the absence of treatment with reducing agents such as 2-ME or DTT, disulfide covalent bonds remain intact, preserving the oligomeric form of the protein. Oligomeric SDS–protein complexes migrate more slowly than their SDS–polypeptide subunits. In addition, nonreduced proteins may not be completely saturated with SDS and hence may not bind the detergent in a constant mass ratio. Moreover, intrachain disulfide bonds constrain the molecular shape, usually in such a way that reduces the Stokes radius of the molecule, thereby reducing the apparent molecular mass, Mr. This makes molecular mass determinations of these molecules by SDS-PAGE less straightforward than analyses of fully denatured polypeptides because it is necessary that both standards and unknown proteins be in similar configurations for valid comparisons.
The use of commercial reagents that have been purified in solution is recommended. When this is not the case and when the purity of the reagents used is not sufficient, a pretreatment is applied. For instance, any solution sufficiently impure to require filtration must also be deionized with a mixed-bed (anion–cation exchange) resin to remove acrylic acid and other charged degradation products. When stored according to recommendations, acrylamide/bisacrylamide solutions and solid persulfate are stable for long periods.
Gel Stock Solutions
30% acrylamide–bisacrylamide solution
Prepare a solution containing 290 g of acrylamide and 10 g of methylenebisacrylamide per liter of water. Filter.
ammonium persulfate solution
Prepare a small quantity of solution having a concentration of 100 g/L of ammonium persulfate. [Note—Ammonium persulfate provides the free radicals that drive polymerization of acrylamide and bisacrylamide. Because ammonium persulfate decomposes rapidly, fresh solutions must be prepared daily.
temed
Use an electrophoresis-grade reagent.
sds solution
This is a 100 g/L solution of electrophoresis-grade SDS.
1.5 m buffer solution
Dissolve 90.8 g of Tris in 400 mL of water. Adjust the pH to 8.8 with hydrochloric acid, and dilute to 500.0 mL with water.
1 m buffer solution
Dissolve 60.6 g of Tris in 400 mL of water. Adjust the pH to 6.8 with hydrochloric acid, and dilute to 500.0 mL with water.
Assembling the Gel-Molding Cassette
Clean the two glass plates (size, e.g., 10 cm × 8 cm), the polytetrafluoroethylene comb, the two spacers, and the silicone rubber tubing (e.g., 0.6 mm diameter × 35 cm length) with mild detergent; rinse extensively with water, followed by dehydrated alcohol; and allow the plates to dry at room temperature. Lubricate the spacers and the tubing with nonsilicone grease. Apply the spacers along each of the two short sides of the glass plate 2 mm away from the edges and 2 mm away from the long side corresponding to the bottom of the gel. Begin to lay the tubing on the glass plate by using one spacer as a guide. Carefully twist the tubing at the bottom of the spacer, and follow the long side of the glass plate. While holding the tubing with one finger along the long side, twist the tubing again and lay it on the second short side of the glass plate, using the spacer as a guide. Place the second glass plate in perfect alignment, and hold the mold together by hand pressure.
Apply two clamps on each of the two short sides of the mold. Carefully apply four clamps on the longer side of the gel mold, thus forming the bottom of the gel mold. Verify that the tubing is running along the edge of the glass plates and has not been extruded while the clamps were placed. The gel mold is now ready for pouring the gel.
Preparation of the Gel
In a discontinuous buffer SDS polyacrylamide gel, it is recommended to pour the resolving gel, let the gel set, and then pour the stacking gel, because the composition of the two gels in acrylamide–bisacrylamide, buffer, and pH are different.
preparation of the resolving gel
In a conical flask, prepare the appropriate volume of solution containing the desired concentration of acrylamide for the resolving gel, using the values given in Table 1. Mix the components in the order shown. Where appropriate, before adding the Ammonium Persulfate Solution and the TEMED, filter the solution if necessary under vacuum through a cellulose acetate membrane (pore diameter: 0.45 µm). Keep the solution under vacuum, while swirling the filtration unit, until no more bubbles are formed in the solution. Add appropriate amounts of Ammonium Persulfate Solution and TEMED, as indicated in Table 1, swirl, and pour immediately into the gap between the two glass plates of the mold. Leave sufficient space for the stacking gel (the length of the teeth of the comb plus 1 cm). Using a tapered glass pipet, carefully overlay the solution with water-saturated isobutanol. Leave the gel in a vertical position at room temperature to allow polymerization.
Table 1. Preparation of the Resolving Gel
|
|
Component Volume (mL) per Gel Mold Volume Below |
|||||||
|---|---|---|---|---|---|---|---|---|
| Solution component |
5 mL |
10 mL |
15 mL |
20 mL |
25 mL |
30 mL |
40 mL |
50 mL |
| 6% Acrylamide |
|
|
|
|
|
|
|
|
| 2.6 |
5.3 |
7.9 |
10.6 |
13.2 |
15.9 |
21.2 |
26.5 |
|
Solution |
1.0 |
2.0 |
3.0 |
4.0 |
5.0 |
6.0 |
8.0 |
10.0 |
| 1.3 |
2.5 |
3.8 |
5.0 |
6.3 |
7.5 |
10.0 |
12.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.004 |
0.008 |
0.012 |
0.016 |
0.02 |
0.024 |
0.032 |
0.04 |
|
| |
|
|
|
|
|
|
|
|
| 8% Acrylamide |
|
|
|
|
|
|
|
|
| 2.3 |
4.6 |
6.9 |
9.3 |
11.5 |
13.9 |
18.5 |
23.2 |
|
Solution |
1.3 |
2.7 |
4.0 |
5.3 |
6.7 |
8.0 |
10.7 |
13.3 |
| 1.3 |
2.5 |
3.8 |
5.0 |
6.3 |
7.5 |
10.0 |
12.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.003 |
0.006 |
0.009 |
0.012 |
0.015 |
0.018 |
0.024 |
0.03 |
|
| |
|
|
|
|
|
|
|
|
| 10% Acrylamide |
|
|
|
|
|
|
|
|
| 1.9 |
4.0 |
5.9 |
7.9 |
9.9 |
11.9 |
15.9 |
19.8 |
|
Solution |
1.7 |
3.3 |
5.0 |
6.7 |
8.3 |
10.0 |
13.3 |
16.7 |
| 1.3 |
2.5 |
3.8 |
5.0 |
6.3 |
7.5 |
10.0 |
12.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.002 |
0.004 |
0.006 |
0.008 |
0.01 |
0.012 |
0.016 |
0.02 |
|
| |
|
|
|
|
|
|
|
|
| 12% Acrylamide |
|
|
|
|
|
|
|
|
| 1.6 |
3.3 |
4.9 |
6.6 |
8.2 |
9.9 |
13.2 |
16.5 |
|
Solution |
2.0 |
4.0 |
6.0 |
8.0 |
10.0 |
12.0 |
16.0 |
20.0 |
| 1.3 |
2.5 |
3.8 |
5.0 |
6.3 |
7.5 |
10.0 |
12.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.002 |
0.004 |
0.006 |
0.008 |
0.01 |
0.012 |
0.016 |
0.02 |
|
| |
|
|
|
|
|
|
|
|
| 14% Acrylamide |
|
|
|
|
|
|
|
|
| 1.4 |
2.7 |
3.9 |
5.3 |
6.6 |
8.0 |
10.6 |
13.8 |
|
Solution |
2.3 |
4.6 |
7.0 |
9.3 |
11.6 |
13.9 |
18.6 |
23.2 |
| 1.2 |
2.5 |
3.6 |
5.0 |
6.3 |
7.5 |
10.0 |
12.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.002 |
0.004 |
0.006 |
0.008 |
0.01 |
0.012 |
0.016 |
0.02 |
|
| |
|
|
|
|
|
|
|
|
| 15% Acrylamide |
|
|
|
|
|
|
|
|
| 1.1 |
2.3 |
3.4 |
4.6 |
5.7 |
6.9 |
9.2 |
11.5 |
|
Solution |
2.5 |
5.0 |
7.5 |
10.0 |
12.5 |
15.0 |
20.0 |
25.0 |
| 1.3 |
2.5 |
3.8 |
5.0 |
6.3 |
7.5 |
10.0 |
12.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.05 |
0.1 |
0.15 |
0.2 |
0.25 |
0.3 |
0.4 |
0.5 |
|
| 0.002 |
0.004 |
0.006 |
0.008 |
0.01 |
0.012 |
0.016 |
0.02 |
|
preparation of the stacking gel
After polymerization is complete (about 30 min), pour off the isobutanol, and wash the top of the gel several times with water to remove the isobutanol overlay and any unpolymerized acrylamide. Drain as much fluid as possible from the top of the gel, and then remove any remaining water with the edge of a paper towel.
In a conical flask, prepare the appropriate volume of solution containing the desired concentration of acrylamide, using the values given in Table 2. Mix the components in the order shown. Where appropriate, before adding the Ammonium Persulfate Solution and the TEMED, filter the solution if necessary under vacuum through a cellulose acetate membrane (pore diameter: 0.45 µm). Keep the solution under vacuum, while swirling the filtration unit, until no more bubbles are formed in the solution. Add appropriate amounts of Ammonium Persulfate Solution and TEMED, as indicated in Table 2. Swirl, and pour immediately into the gap between the two glass plates of the mold directly onto the surface of the polymerized resolving gel. Immediately insert a clean polytetrafluoroethylene comb into the stacking gel solution, being careful to avoid trapping air bubbles. Add more stacking gel solution to fill the spaces of the comb completely. Leave the gel in a vertical position, and allow it to polymerize at room temperature.
Table 2. Preparation of the Stacking Gel
| |
Component Volume (mL) per Gel Mold Volume Below |
|||||||
|---|---|---|---|---|---|---|---|---|
| Solution component |
1 mL |
2 mL |
3 mL |
4 mL |
5 mL |
6 mL |
8 mL |
10 mL |
| Water |
0.68 |
1.4 |
2.1 |
2.7 |
3.4 |
4.1 |
5.5 |
6.8 |
| 30% Acrylamide–Bisacrylamide
Solution |
0.17 |
0.33 |
0.5 |
0.67 |
0.83 |
1.0 |
1.3 |
1.7 |
| 1.0 M Buffer Solution
|
0.13 |
0.25 |
0.38 |
0.5 |
0.63 |
0.75 |
1.0 |
1.25 |
| SDS Solution
|
0.01 |
0.02 |
0.03 |
0.04 |
0.05 |
0.06 |
0.08 |
0.1 |
| Ammonium Persulfate Solution
|
0.01 |
0.02 |
0.03 |
0.04 |
0.05 |
0.06 |
0.08 |
0.1 |
| TEMED
|
0.001 |
0.002 |
0.003 |
0.004 |
0.005 |
0.006 |
0.008 |
0.01 |
Preparation of the Sample
Unless otherwise specified in the specific monograph, the samples can be prepared as follows:
sds-page sample buffer (concentrated)
Dissolve 1.89 g of Tris, 5.0 g of sodium lauryl sulfate, and 50 mg of bromophenol blue in water. Add 25.0 mL of glycerol, and dilute to 100 mL with water. Adjust the pH to 6.8 with hydrochloric acid, and dilute to 125 mL with water.
sds-page sample buffer for reducing conditions (concentrated)
Dissolve 3.78 g of Tris, 10.0 g of SDS, and 100 mg of bromophenol blue in water. Add 50.0 mL of glycerol, and dilute to 200 mL with water. Add 25.0 mL of 2-ME. Adjust to pH 6.8 with hydrochloric acid, and dilute to 250.0 mL with water. Alternatively, DTT can be used as reducing agent instead of 2-ME. In this case prepare the sample buffer as follows: Dissolve 3.78 g of Tris, 10.0 g of SDS, and 100 mg of bromophenol blue in water. Add 50.0 mL of glycerol, and dilute to 200 mL with water. Adjust to pH 6.8 with hydrochloric acid, and dilute to 250.0 mL with water. Immediately before use, add DTT to a final concentration of 100 mM.
sds-page running buffer
Dissolve 151.4 g of Tris, 721.0 g of glycine, and 50.0 g of sodium lauryl sulfate in water, and dilute to 5000 mL with the same solvent. Immediately before use, dilute to 10 times its volume with water, and mix. Measure the pH of the diluted solution. The pH is between 8.1 and 8.8.
sample solution (nonreducing conditions)
Mix equal volumes of: a mixture comprising water plus the preparation or the reference solutions, and SDS-PAGE Sample Buffer (Concentrated).
sample solution (reducing conditions)
Mix equal volumes of: a mixture comprising water plus the preparation or the reference solutions, and SDS-PAGE Sample Buffer for Reducing Conditions (Concentrated) containing 2-ME (or DTT) as the reducing agent.
The concentration prescribed in the monograph can vary depending on the protein and staining method.
Sample treatment: Keep for 5 min in a boiling water bath or in a block heater set at 100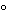 , and then chill. (Note that temperature and time may vary in the monograph because protein cleavage may occur during the heat treatment.)
, and then chill. (Note that temperature and time may vary in the monograph because protein cleavage may occur during the heat treatment.)
Mounting the Gel in the Electrophoresis Apparatus and Electrophoretic Separation
After polymerization is complete (about 30 min), remove the polytetrafluoroethylene comb carefully. Rinse the wells immediately with water or with the SDS-PAGE Running Buffer to remove any unpolymerized acrylamide. If necessary, straighten the teeth of the stacking gel with a blunt hypodermic needle attached to a syringe. Remove the clamps on one short side, carefully pull out the tubing, and replace the clamps. Proceed similarly on the other short side. Remove the tubing from the bottom part of the gel. Mount the gel in the electrophoresis apparatus. Add the electrophoresis buffers to the top and bottom reservoirs. Remove any bubbles that become trapped at the bottom of the gel between the glass plates. This is best done with a bent hypodermic needle attached to a syringe. Never prerun the gel before loading the samples, because this will destroy the discontinuity of the buffer systems. Before loading the sample, carefully rinse each well with SDS-PAGE Running Buffer. Prepare the test and reference solutions in the recommended sample buffer, and treat as specified in the individual monograph. Apply the appropriate volume of each solution to the stacking gel wells.
Start the electrophoresis using the conditions recommended by the equipment manufacturer. Manufacturers of SDS-PAGE equipment may provide gels of different surface area and thickness, and electrophoresis running time and current or voltage may vary in order to achieve optimal separation. Check that the dye front is moving into the resolving gel. When the dye is near the bottom of the gel stop the electrophoresis. Remove the gel assembly from the apparatus, and carefully separate the glass plates. Remove the spacers, cut off and discard the stacking gel, and immediately proceed with staining.
Gradient gels offer some advantages over fixed-concentration gels. Some proteins that co-migrate on fixed-concentration gels can be resolved within gradient gels. During electrophoresis the proteins migrate until the pore size stops further progress, and therefore a stacking effect occurs, resulting in sharper bands. According to Table 3, gradient gels also allow separation of a wider range of protein molecular masses than do single, fixed-concentration gels.
Table 3 gives suggested compositions of the linear gradient, relating the range of acrylamide concentrations to the appropriate protein molecular ranges. Note that other gradient shapes (e.g., concave) can be prepared for specific applications.
Table 3. Acrylamide Gradient Percentages Recommended for
Expected Protein Molecular Weights
Expected Protein Molecular Weights
| Acrylamide (%) |
Protein Range (kDa) |
|---|---|
| 5–15 | 20–250 |
| 5–20 | 10–200 |
| 10–20 | 10–150 |
| 8–20 | 8–150 |
Gradient gels also are used for molecular mass determination and protein purity determination.
Coomassie staining has a protein detection level of approximately 1–10 µg of protein per band. Silver staining is the most sensitive method for staining proteins in gels, and a band containing 10–100 ng can be detected. These figures are considered robust in the context of these gels. Improved sensitivity of one or two orders of magnitude has been reported in the literature.
Coomassie staining responds in a more linear manner than silver staining, but the response and range depend on the protein and development time. Both Coomassie and silver staining can be less reproducible if staining is stopped in a subjective manner, i.e., when the analyst deems the staining satisfactory. Wide dynamic ranges of reference proteins are important to use because they help assess the intra-experimental sensitivity and linearity. All gel-staining steps are done while wearing gloves, at room temperature, with gentle shaking (e.g., on an orbital shaker platform), and using any convenient container.
Staining Reagents
destaining solution
Prepare a mixture of 1 volume of glacial acetic acid, 4 volumes of methanol, and 5 volumes of water.
coomassie staining solution
Prepare a 1.25 g/L solution of acid blue 83 in Destaining Solution. Filter.
fixing solution
To 250 mL of methanol, add 0.27 mL of formaldehyde, and dilute to 500.0 mL with water.
silver nitrate reagent
To a mixture of 3 mL of concentrated ammonia and 40 mL of 1 M sodium hydroxide, add 8 mL of a 200 g/L solution of silver nitrate, dropwise, with stirring. Dilute to 200 mL with water.
developer solution
Dilute 2.5 mL of a 20 g/L solution of citric acid and 0.27 mL of formaldehyde to 500.0 mL with water.
blocking solution
A 10% (v/v) solution of acetic acid.
Coomassie Staining
Immerse the gel in a large excess of Coomassie Staining Solution, and allow to stand for at least 1 h. Remove the staining solution.
Destain the gel with a large excess of Destaining Solution. Change the Destaining Solution several times until the stained protein bands are clearly distinguishable on a clear background. The more thoroughly the gel is destained, the smaller is the amount of protein that can be detected by the method. More rapid destaining can be achieved by including a few grams of anion-exchange resin or a small sponge in the Destaining Solution. [Note—The acid–alcohol solutions used in this procedure do not completely fix proteins in the gel. This can lead to losses of some low-molecular-mass proteins during the staining and destaining of thin gels. Permanent fixation is obtainable by allowing the gel to stand in a mixture of 1 volume of trichloroacetic acid, 4 volumes of methanol, and 5 volumes of water for 1 h before it is immersed in the Coomassie Staining Solution. ]
Silver Staining
Immerse the gel in a large excess of Fixing Solution, and allow it to stand for 1 h. Remove the Fixing Solution, add fresh Fixing Solution, and incubate for at least 1 h or overnight, if convenient. Discard the Fixing Solution, and wash the gel in a large excess of water for 1 h. Soak the gel for 15 min in a 1% (v/v) solution of glutaraldehyde. Wash the gel twice for 15 min in a large excess of water. Soak the gel in fresh Silver Nitrate Reagent for 15 min, in darkness. Wash the gel three times for 5 min in a large excess of water. Immerse the gel for about 1 min in Developer Solution until satisfactory staining has been obtained. Stop the development by incubation in the Blocking Solution for 15 min. Rinse the gel with water.
Depending on the staining method used, gels are treated in a slightly different way. For Coomassie staining, after the destaining step, allow the gel to stand in a 100 g/L solution of glycerol for at least 2 h (overnight incubation is possible). For silver staining, add to the final rinsing a step of 5 min in a 20 g/L solution of glycerol.
Drying of stained SDS polyacrylamide gels is one of the methods to have permanent documentation. This method frequently results in gel cracking during drying between cellulose films.
Immerse two sheets of porous cellulose film in water, and incubate for 5–10 min.
Place one of the sheets on a drying frame. Carefully lift the gel, and place it on the cellulose film. Remove any trapped air bubbles, and pour a few mL of water around the edges of the gel. Place the second sheet on top, and remove any trapped air bubbles. Complete the assembly of the drying frame. Place in an oven or leave at room temperature until dry.
Immediately after the gel has been run, mark the position of the bromophenol blue tracking dye to identify the leading edge of the electrophoretic ion front. This can be done by cutting notches in the edges of the gel or by inserting a needle soaked in India ink into the gel at the dye front. After staining, measure the migration distances of each protein band (markers and unknowns) from the top of the resolving gel. Divide the migration distance of each protein by the distance traveled by the tracking dye. The normalized migration distances are referred to as the relative mobilities of the proteins (relative to the dye front), or RF. Construct a plot of the logarithm of the relative molecular masses (Mr) of the protein standards as a function of the RF values. Unknown molecular masses can be estimated by linear regression analysis (more accurately, by nonlinear regression analysis) or interpolation from the curves of log Mr against RF if the values obtained for the unknown samples are positioned along the approximately linear part of the graph.
Sensitivity also must be validated. A reference protein control corresponding to the desired concentration limit that is run in parallel with the test samples can serve as a system suitability check of the experiment.
When the impurity limit is specified in the individual monograph, analysts should prepare a reference solution corresponding to that level of impurity by diluting the test solution. For example, when the limit is 5%, a reference solution would be a 1:20 dilution of the test solution. No impurity (any band other than the main band) in the electropherogram obtained with the test solution may be more intense than the main band obtained with the reference solution.
Under validated conditions, impurities can be quantified by normalization to the main band, using an integrating densitometer, or by image analysis. 2S (USP38)
2S (USP38)
Auxiliary Information—
Please check for your question in the FAQs before contacting USP.
| Topic/Question | Contact | Expert Committee |
|---|---|---|
| General Chapter | Maura C Kibbey, Ph.D.
Senior Scientific Liaison, Biologics & Biotechnology (301) 230-6309 |
(GCBA2010) General Chapters - Biological Analysis |
USP38–NF33 Page 960
USP38–NF33 Supplement : No. 2 Page 7631
Pharmacopeial Forum: Volume No. 39(2)



