INTRODUCTION
Microorganisms, if detected in drug substances, excipients, water for pharmaceutical use, the manufacturing environment, intermediates, and finished drug products, typically undergo characterization. This may include identification and strain typing, as appropriate. [Note—A Glossary of Terms is provided at the end of this chapter. ] Routine characterization of microorganisms may include the determination of colony morphology, cellular morphology (rods, cocci, cell groupings, modes of sporulation, etc.), Gram reaction or other differential staining techniques, and certain key biochemical reactions (e.g., oxidase, catalase, and coagulase activity) that can be diagnostic. Microbial characterization to this level is sufficient for many risk-assessment purposes in nonsterile pharmaceutical manufacturing operations and in some sterile product manufacturing environments.
In some cases a more definitive identification of the microorganisms yields genus- and species-level identification. Beyond this, available methodologies can perform strain-level identification, which can be useful in an investigation to determine the source of the microorganism. Identification is especially common when organisms are recovered at atypically high rates or in numbers that exceed recommended levels for specific categories of products. Additionally, microbial identification is useful in aseptic processing and is necessary where sterility test positives have occurred and in the assessment of contamination recovered from failed aseptic process simulations, i.e., media fills.
Microbiological identification systems are based on different analytical methodologies, and limitations may be inherent to the method and/or arise from database limitations. Identification is accomplished by matching characteristics (genotypic and/or phenotypic) to an established standard (reference) organism such as a type strain. If a microorganism is not included in the database it will not be identified, so manufacturers should review the breadth of the database of the identification system they plan to use and its applicability to their needs. Users should consider which microbiological identification system(s) is (are) most applicable to their requirements. Bearing in mind both these limitations and the level of identification required (genus, species, strain), users also must select the appropriate technology to use in routine microbiological identification testing.
The need for microbial identification is specifically cited in USP general test chapter Microbiological Examination of Nonsterile Products: Tests for Specified Microorganisms  62
62 . This chapter indicates a requirement for confirmatory identification tests for organisms that grow on selective or diagnostic media and demonstrate defined morphological characteristics. Also, USP general test chapter Sterility Tests
. This chapter indicates a requirement for confirmatory identification tests for organisms that grow on selective or diagnostic media and demonstrate defined morphological characteristics. Also, USP general test chapter Sterility Tests  71
71 allows for invalidation of the test, if after identification of the microorganisms isolated from the test, the growth of this (or these) species may be unequivocally ascribed to faults with respect to the material and/or the technique used in conducting the sterility test procedure. USP general information chapter Microbiological Control and Monitoring of Aseptic Processing Environments
allows for invalidation of the test, if after identification of the microorganisms isolated from the test, the growth of this (or these) species may be unequivocally ascribed to faults with respect to the material and/or the technique used in conducting the sterility test procedure. USP general information chapter Microbiological Control and Monitoring of Aseptic Processing Environments  1116
1116 recommends that microbial isolates be identified at a rate sufficient to support the environmental monitoring program.
recommends that microbial isolates be identified at a rate sufficient to support the environmental monitoring program.
ISOLATION OF PURE CULTURES
The first step in identification is to isolate a pure culture for analysis. This is typically accomplished by successive streaking of the colony of interest in a quadrant pattern on appropriate general microbiological solid media with the objective of obtaining discreet colonies that usually yield pure cultures. This technique also allows phenotypic expression and growth of sufficient inoculum for succeeding identification procedures. Analysts should recognize that expression of the microbial phenotype (i.e., cell size and shape, sporulation, cellular composition, antigenicity, biochemical activity, and sensitivity to antimicrobial agents) may be affected by isolate origins, media selection, and growth conditions (see Table 1). Therefore, the preparatory media for identification and the number of subcultures may affect the results of phenotype identification methods.
Table 1. Phenotypic Characteristics Used in Microbial Taxonomy
| Categories | Characteristics |
|---|---|
| Culture | Colony morphology, colony color, shape and size, pigment production |
| Morphological | Cellular morphology, cell size, cell shape, flagella type, reserve material, Gram reaction, spore and acid-fast staining, mode of sporulation |
| Physiological | Oxygen tolerance, pH range, temperature optimum and range, salinity tolerance |
| Biochemical | Carbon utilization, carbohydrate oxidation or fermentation, enzyme patterns |
| Inhibition | Bile salt-tolerance, antibiotic susceptibility, dye tolerance |
| Serological | Agglutination, fluorescent antibody |
| Chemo-taxonomic | Fatty acid profile, microbial toxins, whole cell composition |
| Ecological | Origin of the organism |
In contrast, the microbial genotype generally is well conserved and unaffected by cultural conditions. Therefore, once the isolation of a pure, monoclonal colony is assured, the microorganism may be analyzed without concern over the most recent growth media or the viability of the isolate. Table 2 lists genotypic characteristics that can be determined.
Table 2. Genotypic/Phylogenetic Characteristics That Can Be Used in Microbial Taxonomy
| Categories | Characteristics |
| Genotypic | DNA base ratio (G + C content), restriction fragment patterns, and DNA probes |
| Phylogenetic | DNA–DNA hybridization, and 16S and 23S rRNA sequences |
Bacterial taxonomy as described in Bergey's Manual of Systematic Bacteriology (Bergey's Manual)1 is at present accomplished by comparative analysis of genetic material. When the DNA from an unknown organism is compared to the DNA from a known organism, the degree of relatedness can be determined. Genotypic identification (Table 2) is accomplished through the use of DNA hybridization, restriction fragment pattern comparisons, and/or DNA probes. For example, greater than 70% relatedness with DNA–DNA hybridization indicates the organisms are the same species. Phylogenetic analysis (Table 2) is typically performed by comparing the base sequence of a portion of the 16S ribosomal RNA gene for bacteria, or the 23S ribosomal RNA gene for fungi. Polymerase chain reaction (PCR) is used to amplify these genes, and the amplified region is then isolated and base sequenced using an electrophoretic or dideoxy chain termination method. Comparisons can be made using validated proprietary databases or those that are publicly available. [Caution: Publicly available databases may not be validated].
PRIMARY SCREENING AND CHARACTERIZATION
Microorganisms isolated on compendial media from samples of pharmaceutical ingredients, water for pharmaceutical use, the manufacturing environment, intermediates, and finished products may be physiologically stressed. The microorganisms will pass from a metabolic state suitable for survival under adverse ambient conditions to culture conditions that are far richer nutritionally and are at an optimal incubation temperature. This transition can be managed by careful handling of the isolates. In preparation for identification, individual representative colonies from the primary isolation media are streaked for monoclonal colonies onto solid media as described above. The first step is to determine the Gram reaction, cellular morphology, and in some cases diagnostic biochemical reactions of the bacteria isolates. This is a critical step for many phenotypic identification schemes. If the wrong characteristics are assigned to an isolate, subsequent testing may be conducted using the wrong microbial identification kit, resulting in an incorrect result. Several common preliminary screening tests are described below.
Gram Staining
Gram staining methods include the four-step method: crystal violet (primary stain), iodine (mordant), alcohol or alcohol–acetone (decolorizer), and safranin (counterstain). In the three-step method the decolorization and counterstaining steps are combined. Under optimal conditions, Gram-positive organisms retain the crystal violet stain and appear blue violet. Gram-negative organisms lose the crystal violet stain, so they contain only the counterstain safranin and appear red. Some bacteria may be Gram-variable. Common pitfalls in this method are that heat fixation may cause Gram-positive cells to stain Gram-negative, and older cultures may give Gram-variable reactions. Using too much decolorizer could result in a false Gram-negative result, and not using enough decolorizer may yield a false Gram-positive result. One variation that has advantages in some situations is to perform a methanol, rather than heat, fixation of the bacterial smear. In some cases alcohol fixation may give more consistent Gram stain results. In either method a Gram-positive and a Gram-negative control should be included to allow identification of errors in staining. Because the Gram-staining reaction must be read under a microscope, cellular morphology can be simultaneously ascertained.
Spore Staining
Spore staining can be accomplished using a malachite green stain for bacterial spores. A positive control should be included to allow identification of errors in spore staining.
Biochemical Screening
Key biochemical screening tests include (1) the oxidase test to separate Gram-negative, rod-shaped bacteria into nonfermenters (oxidase positive) and enteric (oxidase negative) bacteria, (2) the catalase test to separate Staphylococci (catalase positive) from Streptococci (catalase negative), and (3) the coagulase test to separate Staphylococci into coagulase negative (presumptively nonpathogenic) and coagulase positive (more likely pathogenic) Staphylococci.
For many types of investigations and routine surveying of manufacturing environmental bioburden, these few tests can provide sufficient information for ongoing evaluation. However, when circumstances dictate greater in-depth assessment, identification to the genus, species, or strain level can yield valuable insights about the nature and source of environmental bioburden. Also, microbial identification to the species and even strain level can be critical in assessing and mitigating risk from microbial contamination.
MICROBIAL IDENTIFICATION BY PHENOTYPIC METHODS
Phenotypic methods use expressed gene products to distinguish among different microorganisms. Generally, these require a relatively large number of cells in pure, monoclonal culture. Recovery and growth methods for microbial enumeration and identification are limited by the length of incubation and the fact that many organisms present in the environment are not recovered by general microbiological growth media. Additionally, freshly isolated, stressed microorganisms by subculture from primary recovery may not result in a full expression of phenotypic properties. However, methods based on carbon utilization and biochemical reaction, as well as fatty acid profiles by gas–liquid chromatography and whole-cell composition by MALDI–TOF mass spectrometry, are always based on inocula development for a specific identification system. These systems rely on specified culture media and incubation conditions to achieve consistent identification. Phenotypic microbial identification methods are successfully used in food, water, clinical, and pharmaceutical microbiological testing laboratories.2 Phenotypic microbial identification methods provide information that enables microbiologists to make informed decisions regarding product risk and to recognize changes in environmental microflora. In many quality control investigations, phenotypic identification alone is sufficient and will enable scientists to conduct a thorough investigation and to recommend appropriate corrective actions as needed.
MICROBIAL IDENTIFICATION BY GENOTYPIC METHODS
Genotypic microbial identification methods are theoretically more reliable because nucleic acid sequences are highly conserved in most microbial species. Applicable genotypic methods include DNA–DNA hybridization, PCR, 16S and 23S rRNA sequencing, multilocus sequence typing (MLST), pyrosequencing, DNA probes, and analytical ribotyping. These methods can be technically challenging for microbiologists. They also require more expensive analytical equipment and supplies. Often these analyses are conducted by contract laboratories, government laboratories, universities, research institutes, or specialized laboratories within industrial firms. Therefore, the use of genotypic identification methods is typically limited to critical microbiological investigations such as product failure investigations. Further, if strain-level identification is done in the course of an investigation, analysts must ensure that the method is appropriate.
DNA sequencing of the first 500 base pairs of the 16S rRNA sequence is useful for identification to the species level but may not provide sufficient power to resolve among closely related species or strains of the same species. In contrast, Southern hybridization of restriction endonuclease digests is powerful and can be effective in demonstrating differences between two strains. If the banding patterns appear identical, this shows only that restriction endonuclease has similar cleavage sites in that region of the two organisms. Demonstration that the two organisms are the same should include two or more different restriction endonuclease digests, each of which yields bands in the area of interest. All bands from the two organisms must be identical.
In contrast to microbial identification, nucleic acid-based methods can be used to screen for specific microorganisms. The steps associated with this activity are sample collection, nucleic acid extraction, target amplification, hybridization, and detection. The problem of amplifying DNA from nonviable bacterial cells can be overcome by using reverse transcription to convert rRNA that is transitional, hence related to viability, to DNA for PCR amplification. Issues include the detection of microbial variants, limits of detection, matrix effects, positive cutoff verification, instrument and system carry-over, diagnostic accuracy, and reproducibility.
VERIFICATION OF MICROBIAL IDENTIFICATION METHODS
Microbial identification tests include serological tests, chemical reagents, reference organisms, and instrumentation. The verification of an identification test system can include one of the following: (1) using an existing system for parallel testing of microbial isolates obtained from routine testing (the number of isolates tested may be as high as 50, and any discrepancies in identification can be arbitrated using a referee method); (2) testing 12–15 known representative stock cultures of different commonly isolated species for a total of 50 tests; or (3) confirming that 20–50 organism identifications, including 15–20 different species, agree with the results of a reference laboratory testing of split sample.3 In each case the appropriate quality control organisms, as recommended by the supplier and the compendia, should be included in the verification process.
With identification systems, verification of the identity of the species should be evaluated and the level of agreement should be considered. Typically greater than 90% agreement can be achieved with samples of microorganisms that are appropriate for the identification system. Groups of organisms that are challenging to identify (e.g., nonfermenting bacteria, corynebacteria, and coagulase-negative Staphylococci) may be included, when appropriate, in the verification process but may yield lower levels of agreement.
The hierarchy of microbial identification errors in descending order of impact is (1) misidentification to genera, (2) misidentification to species, and (3) no identification. Misidentification could lead to inappropriate corrective and preventive actions and product disposition.
A microbial identification system may not be able to identify an isolate because the organism is not included in the database, the system parameters are not sufficiently comprehensive to identify the organism, the isolate may be nonreactive in the system, or the species may not have been taxonomically described. Such isolates can be sent to the supplier of the microbial identification system for additional study and, if appropriate, added to the database. Alternatively, genotypic identification tests can be conducted, and the species can be added to an in-house database. Misidentification is more difficult to determine, but any microbial identification should be reviewed for reasonableness in terms of the microorganism’s morphology, physiological requirements, and source of isolation. Organisms identified only to genus may be common for the numerous nonpathogenic species of Staphylococcus, Corynebacterium (and other small pleomorphic Gram-positive rods), and Micrococcus.
The most important verification tests are accuracy and reproducibility. These measurements can be defined as follows:
Accuracy % = (Number of correct results/Total number of results) × 100
Reproducibility % = (Number of correct results in agreement/Total number of results) × 100
The user should establish suitable acceptance criteria for accuracy and reproducibility, taking into account method capability.
Other measurements are sensitivity, specificity, and positive and negative predictive value. These measurements are best illustrated by an example. A clinical microbiology laboratory compared the frequency of isolation of a DNA hybridization probe to a culture method for the sexually transmitted bacterium Neisseria gonorrhaeae.3 The frequency of isolation from clinical specimens was historically 10%. The lab ran 100 split samples, and the results are presented in Table 3.
Table 3. Comparison of the Distribution of Negative and Positive Results for the DNA Probe and Culture Methods
| Culture Results | ||
|---|---|---|
| DNA Probe Results | Positive | Negative |
| Positive | 9 | 2 |
| Negative | 1 | 88 |
Sensitivity = [9/(9 + 1)] × 100 = 90%
Specificity = [88/(88 + 2)] × 100 = 97.7%
Positive Predictive Value = [9/(9 + 2)] × 100 = 81.8%
Negative Predictive Value = [88/(88 + 1)] × 100 = 98.9%
Note that the positive predictive value (PPV) is not intrinsic to the test; it also depends on the prevalence of the microorganism in clinical samples. PPV is directly proportional to the prevalence of the disease or condition. In this example, if the group of people tested had included a higher proportion of people with infection, then the PPV would probably be higher and the negative predictive value (NPV) lower. If all persons in the group had infection, the PPV would be 100% and the NPV 0%. The mathematical derivation of these functions is outlined in Table 4.
Table 4. A Two-Row by Two-Column Contingency Table with Respect to the Reference Culture Method and the Alternate PCR Method (After ISO 5725-1 and 5725-2 2004)*
| PCR | |||
|---|---|---|---|
| Culture | Positive | Negative | Sum |
| Positive | a True Positive |
b False Negative |
a + b |
| Negative | c False Positive |
d True Negative |
c + d |
| Sum | a + c | b + d | |
|
*
ISO 5725-1:1994 Accuracy (trueness and precision) of measurement methods and results—Part 1: General principles and definitions and ISO 5725-2:1994 Accuracy (trueness and precision) of measurement methods and results—Part 2: Basic methods for the determination of repeatability and reproducibility of standard measurement methods.
|
|||
Inclusivity (%) = [a/(a + b)] × 100
Exclusivity (%) = [d/(c + d)] × 100
Positive Predictivity (%) = [a/(a + c)] × 100
Negative Predictivity (%) = [d/(b + d)] × 100
Analytical Accuracy (%) = [(a + d)/(a + b + c + d)] × 100
Kappa Index = 2(ad
Phylogenetic Considerations
The second edition of Bergey's Manual represented a major departure from the first edition, and also from the eighth and ninth editions of the Manual of Determinative Bacteriology. The organization of content in Bergey's Manual follows a phylogenetic framework, based on analysis of the nucleotide sequence of the ribosomal small subunit 16S RNA, rather than a phenotypic structure.
Phylogenetic trees or dendrograms show the closest genetically related organisms. The application of this technology has resulted in taxonomic revisions and the renaming of some well-known microorganisms; e.g., the fungus A. niger ATCC 16404 was renamed A. brasiliensis. In general, organisms with relatedness less than or equal to 97% are considered different genera and those with relatedness less than or equal to 99% are considered different species,4 but there are many exceptions to this generalization.
Differences in genotype and phenotype are relatively uncommon, e.g., same or very similar genotype shared by microorganisms with different phenotypes, similar phenotypes but different genotypes, and microorganisms that are genotypically too distant to be the same species or genus. The concept of polyphasic taxonomy5 that refers to assembly and use of many levels of information, e.g., microbial characterization, phenotypic and genotypic data, and origin of the microorganisms, can be successfully applied to microbial identification. This avoids decisions made solely using genotypic data that make no sense when the microbial characteristics, testing history, and source of isolation are considered.
GLOSSARY OF TERMS
Microbial classification:
The arrangement of microorganisms into taxonomic groups based on their similarities and relationships.
Microbial identification:
The determination of which broad group (e.g., bacteria, yeast, or mold) or narrow group (e.g., genus and/or species) to which a laboratory isolate belongs.
Microbial characterization:
The use of colony growth, cellular morphology, differential staining, and key diagnostic features to characterize a laboratory isolate for trending and investigative purposes without identification, e.g,. nonpathogenic Staphylococci.
Mol %GC:
The molecular percentage of guanine–cytosine range within the chromosomal DNA. [Note—The %GC + %AT = 100%. ]
Phylogenetic species:
A species consisting of many strains including the type of strain that shares at least 70% total genome DNA–DNA hybridization and less than 5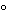 DTm (difference in melting point of the hybrid).
DTm (difference in melting point of the hybrid).
Polyphasic taxonomy:
Taxonomy that assembles and assimilates many levels of information from molecular, physiological, morphological, serological, or ecological sources to classify a microorganism.
Relatedness:
The extent of relationship or similarity of two (or more) organisms on a Phylogenetic Tree or a Dendrogram.
rRNA Sequence:
The DNA sequences that encode rRNA used in protein synthesis are highly conserved among microorganisms of a common ancestry. They are used to determine the phylogenetic distance between organisms and are useful in microbial taxonomy and identification.
Strain:
A specific isolate of a species that is maintained in pure culture and is characterized. The type strain is representative of the species that provides a reference for the species based on its historic isolation, characterization, and deposition in recognized culture collections.
Strain typing:
Strain typing is an integral part of epidemiological investigations in clinical and public health microbiology. Methods including pulsed-field gel electrophoresis, riboprinting, arbitrarily primed polymerized chain reaction, and whole genome ordered restriction or optical mapping can be used to demonstrate that microbial species are the same strain and most likely are from a common source.
1
Bergey's Manual of Systematic Bacteriology, 2nd Edition, 2003.
2
O'Hara, C.M., M.P. Weinstein, and J.M. Miller. Manual and automated systems for detection and identification of microorganisms. ASM Manual of Clinical Microbiology, 8th Edition, 2003.
3
Cumitech 31. Verification and Validation of Procedures in the Clinical Microbiology Laboratory. Elder, B.L., S.A. Hansen, J.A. Kellogg, F.J. Marsik, and R.J. Zabransky, ASM, February 1997.
4
J.E. Clarridge III. The Impact of 16S rRNA Gene Sequencing Analysis for Identification of Bacteria on Clinical Microbiology and Infectious Diseases, Clin. Microbiol. Rev. 17 (2) 840–862, 2004.
5
Gillis, M., P. Vandamme, P. De Vos, J. Swings and K. Kersters. Polyphasic Taxonomy. Bergey's Manual of Systematic Bacteriology, 2nd Edition, 2003.
Auxiliary Information—
Please check for your question in the FAQs before contacting USP.
| Topic/Question | Contact | Expert Committee |
|---|---|---|
| General Chapter | Radhakrishna S Tirumalai, Ph.D.
Principal Scientific Liaison (301) 816-8339 |
(GCM2010) General Chapters - Microbiology |
USP38–NF33 Page 1180
Pharmacopeial Forum: Volume No. 36(6) Page 1675



